The Rhinovirus subviral a-particle exposes 3'-terminal sequences of its genomic RNA
- PMID: 24672023
- PMCID: PMC4093901
- DOI: 10.1128/JVI.00539-14
The Rhinovirus subviral a-particle exposes 3'-terminal sequences of its genomic RNA
Abstract
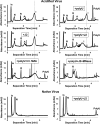
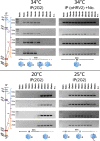
Enteroviruses, which represent a large genus within the family Picornaviridae, undergo important conformational modifications during infection of the host cell. Once internalized by receptor-mediated endocytosis, receptor binding and/or the acidic endosomal environment triggers the native virion to expand and convert into the subviral (altered) A-particle. The A-particle is lacking the internal capsid protein VP4 and exposes N-terminal amphipathic sequences of VP1, allowing for its direct interaction with a lipid bilayer. The genomic single-stranded (+)RNA then exits through a hole close to a 2-fold axis of icosahedral symmetry and passes through a pore in the endosomal membrane into the cytosol, leaving behind the empty shell. We demonstrate that in vitro acidification of a prototype of the minor receptor group of common cold viruses, human rhinovirus A2 (HRV-A2), also results in egress of the poly(A) tail of the RNA from the A-particle, along with adjacent nucleotides totaling ∼700 bases. However, even after hours of incubation at pH 5.2, 5'-proximal sequences remain inside the capsid. In contrast, the entire RNA genome is released within minutes of exposure to the acidic endosomal environment in vivo. This finding suggests that the exposed 3'-poly(A) tail facilitates the positioning of the RNA exit site onto the putative channel in the lipid bilayer, thereby preventing the egress of viral RNA into the endosomal lumen, where it may be degraded.
Importance: For host cell infection, a virus transfers its genome from within the protective capsid into the cytosol; this requires modifications of the viral shell. In common cold viruses, exit of the RNA genome is prepared by the acidic environment in endosomes converting the native virion into the subviral A-particle. We demonstrate that acidification in vitro results in RNA exit starting from the 3'-terminal poly(A). However, the process halts as soon as about 700 bases have left the viral shell. Conversely, inside the cell, RNA egress completes in about 2 min. This suggests the existence of cellular uncoating facilitators.
Figures



Similar articles
-
Viral uncoating is directional: exit of the genomic RNA in a common cold virus starts with the poly-(A) tail at the 3'-end.PLoS Pathog. 2013;9(4):e1003270. doi: 10.1371/journal.ppat.1003270. Epub 2013 Apr 4. PLoS Pathog. 2013. PMID: 23592991 Free PMC article.
-
Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle.Proc Natl Acad Sci U S A. 2013 Dec 10;110(50):20063-8. doi: 10.1073/pnas.1312128110. Epub 2013 Nov 25. Proc Natl Acad Sci U S A. 2013. PMID: 24277846 Free PMC article.
-
Productive entry pathways of human rhinoviruses.Adv Virol. 2012;2012:826301. doi: 10.1155/2012/826301. Epub 2012 Nov 26. Adv Virol. 2012. PMID: 23227049 Free PMC article.
-
Uncoating of human rhinoviruses.Rev Med Virol. 2010 Sep;20(5):281-97. doi: 10.1002/rmv.654. Rev Med Virol. 2010. PMID: 20629045 Review.
-
Proposals for the classification of human rhinovirus species C into genotypically assigned types.J Gen Virol. 2010 Oct;91(Pt 10):2409-19. doi: 10.1099/vir.0.023994-0. Epub 2010 Jul 7. J Gen Virol. 2010. PMID: 20610666 Review.
Cited by
-
Enterovirus particles expel capsid pentamers to enable genome release.Nat Commun. 2019 Mar 8;10(1):1138. doi: 10.1038/s41467-019-09132-x. Nat Commun. 2019. PMID: 30850609 Free PMC article.
-
Stabilization of the Quadruplex-Forming G-Rich Sequences in the Rhinovirus Genome Inhibits Uncoating-Role of Na+ and K.Viruses. 2023 Apr 19;15(4):1003. doi: 10.3390/v15041003. Viruses. 2023. PMID: 37112983 Free PMC article.
-
Rhinovirus Inhibitors: Including a New Target, the Viral RNA.Viruses. 2021 Sep 7;13(9):1784. doi: 10.3390/v13091784. Viruses. 2021. PMID: 34578365 Free PMC article. Review.
-
Impairing the function of MLCK, myosin Va or myosin Vb disrupts Rhinovirus B14 replication.Sci Rep. 2017 Dec 7;7(1):17153. doi: 10.1038/s41598-017-17501-z. Sci Rep. 2017. PMID: 29215055 Free PMC article.
-
Developing a Novel Gene-Delivery Vector System Using the Recombinant Fusion Protein of Pseudomonas Exotoxin A and Hyperthermophilic Archaeal Histone HPhA.PLoS One. 2015 Nov 10;10(11):e0142558. doi: 10.1371/journal.pone.0142558. eCollection 2015. PLoS One. 2015. PMID: 26556098 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

