NTPDASE4 gene products cooperate with the adenovirus E4orf4 protein through PP2A-dependent and -independent mechanisms and contribute to induction of cell death
- PMID: 24672025
- PMCID: PMC4093884
- DOI: 10.1128/JVI.00381-14
NTPDASE4 gene products cooperate with the adenovirus E4orf4 protein through PP2A-dependent and -independent mechanisms and contribute to induction of cell death
Abstract
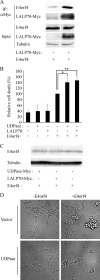
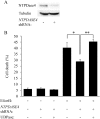
The adenovirus E4orf4 protein induces nonclassical apoptosis in mammalian cells through at least two complementing pathways regulated by the interactions of E4orf4 with protein phosphatase 2A (PP2A) and Src kinases. In Saccharomyces cerevisiae cells, which do not express Src, E4orf4 induces PP2A-dependent toxicity. The yeast Golgi apyrase Ynd1 was found to contribute to E4orf4-mediated toxicity and to interact with the PP2A-B55α regulatory subunit. In addition, a mammalian Ynd1 orthologue, the NTPDASE4 gene product Golgi UDPase, was shown to physically interact with E4orf4. Here we report that knockdown of NTPDASE4 suppressed E4orf4-induced cell death. Conversely, overexpression of the NTPDASE4 gene products Golgi UDPase and LALP70 enhanced E4orf4-induced cell killing. We found that similarly to results obtained in yeast, the apyrase activity of mammalian UDPase was not required for its contribution to E4orf4-induced toxicity. The interaction between E4orf4 and UDPase had two consequences: a PP2A-dependent one, resulting in increased UDPase levels, and a PP2A-independent outcome that led to dissociation of large UDPase-containing protein complexes. The present report extends our findings in yeast to E4orf4-mediated death of mammalian cells, and combined with previous results, it suggests that the E4orf4-NTPDase4 pathway, partly in association with PP2A, may provide an alternative mechanism for the E4orf4-Src pathway to contribute to the cytoplasmic death function of E4orf4.
Importance: The adenovirus E4orf4 protein contributes to regulation of the progression of virus infection from the early to the late phase, and when expressed alone, it induces a unique caspase-independent programmed cell death which is more efficient in cancer cells than in normal cells. The interactions of E4orf4 with cellular proteins that mediate its functions, such as PP2A and Src kinases, are highly conserved in evolution. The results presented here reveal that the NTPDASE4 gene product Golgi UDPase, first discovered to contribute to E4orf4 toxicity in Saccharomyces cerevisiae, associates with E4orf4 and plays a role in induction of cell death in mammalian cells. Details of the functional interaction between E4orf4, PP2A, and the UDPase are described. Identification of the evolutionarily conserved mechanisms underlying E4orf4 activity will increase our understanding of the interactions between the virus and the host cell and will contribute to our grasp of the unique mode of E4orf4-induced cell death.
Figures






Similar articles
-
Mechanisms of cancer cell killing by the adenovirus E4orf4 protein.Viruses. 2015 May 7;7(5):2334-57. doi: 10.3390/v7052334. Viruses. 2015. PMID: 25961489 Free PMC article. Review.
-
YND1 interacts with CDC55 and is a novel mediator of E4orf4-induced toxicity.J Biol Chem. 2005 Dec 16;280(50):41270-7. doi: 10.1074/jbc.M507281200. Epub 2005 Oct 14. J Biol Chem. 2005. PMID: 16227198
-
The cytosolic tail of the Golgi apyrase Ynd1 mediates E4orf4-induced toxicity in Saccharomyces cerevisiae.PLoS One. 2010 Nov 22;5(11):e15539. doi: 10.1371/journal.pone.0015539. PLoS One. 2010. PMID: 21124936 Free PMC article.
-
Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A.Oncogene. 2001 Aug 30;20(38):5279-90. doi: 10.1038/sj.onc.1204693. Oncogene. 2001. PMID: 11536041
-
Biology of the adenovirus E4orf4 protein: from virus infection to cancer cell death.FEBS Lett. 2020 Jun;594(12):1891-1917. doi: 10.1002/1873-3468.13704. Epub 2019 Dec 12. FEBS Lett. 2020. PMID: 31792953 Review.
Cited by
-
The Human Adenovirus Type 5 E4orf4 Protein Targets Two Phosphatase Regulators of the Hippo Signaling Pathway.J Virol. 2015 Sep;89(17):8855-70. doi: 10.1128/JVI.03710-14. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085163 Free PMC article.
-
Viral genes as oncolytic agents for cancer therapy.Cell Mol Life Sci. 2015 Mar;72(6):1073-94. doi: 10.1007/s00018-014-1782-1. Epub 2014 Nov 19. Cell Mol Life Sci. 2015. PMID: 25408521 Free PMC article. Review.
-
Mechanisms of cancer cell killing by the adenovirus E4orf4 protein.Viruses. 2015 May 7;7(5):2334-57. doi: 10.3390/v7052334. Viruses. 2015. PMID: 25961489 Free PMC article. Review.
-
The adenoviral E4orf4 protein: A multifunctional protein serving as a guide for treating cancer, a multifactorial disease.Tumour Virus Res. 2025 Jun;19:200303. doi: 10.1016/j.tvr.2024.200303. Epub 2024 Dec 15. Tumour Virus Res. 2025. PMID: 39681196 Free PMC article.
-
An Interaction with PARP-1 and Inhibition of Parylation Contribute to Attenuation of DNA Damage Signaling by the Adenovirus E4orf4 Protein.J Virol. 2019 Sep 12;93(19):e02253-18. doi: 10.1128/JVI.02253-18. Print 2019 Oct 1. J Virol. 2019. PMID: 31315986 Free PMC article.
References
-
- Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins, Nature 393:185–187 - PubMed
-
- Mannervik M, Fan S, Strom AC, Helin K, Akusjarvi G. 1999. Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256:313–321. 10.1006/viro.1999.9663 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

