A unique N-terminal sequence in the Carnation Italian ringspot virus p36 replicase-associated protein interacts with the host cell ESCRT-I component Vps23
- PMID: 24672030
- PMCID: PMC4093892
- DOI: 10.1128/JVI.03840-13
A unique N-terminal sequence in the Carnation Italian ringspot virus p36 replicase-associated protein interacts with the host cell ESCRT-I component Vps23
Abstract
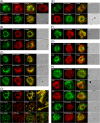
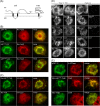
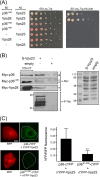
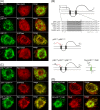
Like most positive-strand RNA viruses, infection by plant tombusviruses results in extensive rearrangement of specific host cell organelle membranes that serve as the sites of viral replication. The tombusvirus Tomato bushy stunt virus (TBSV) replicates within spherules derived from the peroxisomal boundary membrane, a process that involves the coordinated action of various viral and cellular factors, including constituents of the endosomal sorting complex required for transport (ESCRT). ESCRT is comprised of a series of protein subcomplexes (i.e., ESCRT-0 -I, -II, and -III) that normally participate in late endosome biogenesis and some of which are also hijacked by certain enveloped retroviruses (e.g., HIV) for viral budding from the plasma membrane. Here we show that the replication of Carnation Italian ringspot virus (CIRV), a tombusvirus that replicates at mitochondrial membranes also relies on ESCRT. In plant cells, CIRV recruits the ESCRT-I protein, Vps23, to mitochondria through an interaction that involves a unique region in the N terminus of the p36 replicase-associated protein that is not conserved in TBSV or other peroxisome-targeted tombusviruses. The interaction between p36 and Vps23 also involves the Vps23 C-terminal steadiness box domain and not its N-terminal ubiquitin E2 variant domain, which in the case of TBSV (and enveloped retroviruses) mediates the interaction with ESCRT. Overall, these results provide evidence that CIRV uses a unique N-terminal sequence for the recruitment of Vps23 that is distinct from those used by TBSV and certain mammalian viruses for ESCRT recruitment. Characterization of this novel interaction with Vps23 contributes to our understanding of how CIRV may have evolved to exploit key differences in the plant ESCRT machinery.
Importance: Positive-strand RNA viruses replicate their genomes in association with specific host cell membranes. To accomplish this, cellular components responsible for membrane biogenesis and modeling are appropriated by viral proteins and redirected to assemble membrane-bound viral replicase complexes. The diverse pathways leading to the formation of these replication structures are poorly understood. We have determined that the cellular ESCRT system that is normally responsible for mediating late endosome biogenesis is also involved in the replication of the tombusvirus Carnation Italian ringspot virus (CIRV) at mitochondria. Notably, CIRV recruits ESCRT to the mitochondrial outer membrane via an interaction between a unique motif in the viral protein p36 and the ESCRT component Vps23. Our findings provide new insights into tombusvirus replication and the virus-induced remodeling of plant intracellular membranes, as well as normal ESCRT assembly in plants.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

