Substitution of herpes simplex virus 1 entry glycoproteins with those of saimiriine herpesvirus 1 reveals a gD-gH/gL functional interaction and a region within the gD profusion domain that is critical for fusion
- PMID: 24672037
- PMCID: PMC4093879
- DOI: 10.1128/JVI.00465-14
Substitution of herpes simplex virus 1 entry glycoproteins with those of saimiriine herpesvirus 1 reveals a gD-gH/gL functional interaction and a region within the gD profusion domain that is critical for fusion
Abstract
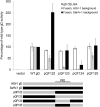
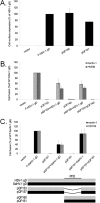
To gain insight into the mechanism of herpesvirus entry into cells, the four glycoproteins that are necessary for herpes simplex virus (HSV) fusion were cloned from the saimiriine herpesvirus 1 (SaHV-1) genome, a primate member of the alphaherpesvirus family. Cell-cell fusion assays indicate that SaHV-1 entry glycoproteins function with the previously identified alphaherpesvirus entry receptors nectin-1 and CD155 but not with herpesvirus entry mediator (HVEM) or paired immunoglobulin-like type 2 receptor alpha (PILRα). Replacement of HSV-1 gD with the SaHV-1 gD homolog resulted in a complete loss of fusion function when coexpressed with HSV-1 gB and gH/gL. HSV-1 gD was also unable to substitute for SaHV-1 gD when coexpressed with SaHV-1 gB and gH/gL. Similarly, the gH/gL heterodimers from HSV-1 and SaHV-1 were not interchangeable. In contrast, both the HSV-1 and SaHV-1 gB homologs retained function in a heterotypic context. These results suggest that an essential interaction between homotypic gD and gH/gL occurs during both HSV-1 and SaHV-1 entry. To map the site of this homotypic interaction, we created a series of gD chimeras, focusing on the "profusion domain" (PFD) that consists of HSV-1 gD residues 261 to 305 or SaHV-1 gD residues 264 to 307. We identified a seven-amino-acid stretch (264 RTLPPPK 270) at the N terminus of the SaHV-1 gD PFD that contributes to homotypic fusion. Finally, we found that the gD receptor-binding region and PFD cannot function independently but that both can inhibit the function of wild-type gD.
Importance: The herpesvirus entry machinery requires the concerted action of at least four glycoproteins; however, details of the interactions among these glycoproteins are not well understood. Like HSV-1, SaHV-1 belongs to the alphaherpesvirus subfamily. Using cell-cell fusion experiments, we found that SaHV-1 uses the entry receptors nectin-1 and CD155 but not HVEM or PILRα. By swapping the entry glycoproteins between HSV-1 and SaHV-1, we revealed a functional interaction between gD and gH/gL. To examine the homotypic interaction site on gD, we evaluated the function of a panel of HSV-1/SaHV-1 gD chimeras and identified a small region in the SaHV-1 gD profusion domain that is critical for SaHV-1 fusion. This study contributes to our understanding of the molecular mechanisms of herpesvirus entry and membrane fusion.
Figures








Similar articles
-
Multiple Sites on Glycoprotein H (gH) Functionally Interact with the gB Fusion Protein to Promote Fusion during Herpes Simplex Virus (HSV) Entry.mBio. 2023 Feb 28;14(1):e0336822. doi: 10.1128/mbio.03368-22. Epub 2023 Jan 11. mBio. 2023. PMID: 36629412 Free PMC article.
-
Two Sides to Every Story: Herpes Simplex Type-1 Viral Glycoproteins gB, gD, gH/gL, gK, and Cellular Receptors Function as Key Players in Membrane Fusion.Viruses. 2021 Sep 16;13(9):1849. doi: 10.3390/v13091849. Viruses. 2021. PMID: 34578430 Free PMC article. Review.
-
A Functional Interaction between Herpes Simplex Virus 1 Glycoprotein gH/gL Domains I and II and gD Is Defined by Using Alphaherpesvirus gH and gL Chimeras.J Virol. 2015 Jul;89(14):7159-69. doi: 10.1128/JVI.00740-15. Epub 2015 Apr 29. J Virol. 2015. PMID: 25926636 Free PMC article.
-
Herpes B virus utilizes human nectin-1 but not HVEM or PILRα for cell-cell fusion and virus entry.J Virol. 2012 Apr;86(8):4468-76. doi: 10.1128/JVI.00041-12. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345445 Free PMC article.
-
The multipartite system that mediates entry of herpes simplex virus into the cell.Rev Med Virol. 2007 Sep-Oct;17(5):313-26. doi: 10.1002/rmv.546. Rev Med Virol. 2007. PMID: 17573668 Review.
Cited by
-
Extracellular Vesicles Released by Bovine Alphaherpesvirus 1-Infected A549 Cells May Limit Subsequent Infections of the Progeny Virus.Int J Mol Sci. 2025 Jun 26;26(13):6181. doi: 10.3390/ijms26136181. Int J Mol Sci. 2025. PMID: 40649956 Free PMC article.
-
Multiple Sites on Glycoprotein H (gH) Functionally Interact with the gB Fusion Protein to Promote Fusion during Herpes Simplex Virus (HSV) Entry.mBio. 2023 Feb 28;14(1):e0336822. doi: 10.1128/mbio.03368-22. Epub 2023 Jan 11. mBio. 2023. PMID: 36629412 Free PMC article.
-
Receptor Binding-Induced Conformational Changes in Herpes Simplex Virus Glycoprotein D Permit Interaction with the gH/gL Complex to Activate Fusion.Viruses. 2023 Mar 30;15(4):895. doi: 10.3390/v15040895. Viruses. 2023. PMID: 37112875 Free PMC article.
-
Regulation of Herpes Simplex Virus Glycoprotein-Induced Cascade of Events Governing Cell-Cell Fusion.J Virol. 2016 Nov 14;90(23):10535-10544. doi: 10.1128/JVI.01501-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27630245 Free PMC article.
-
Two Sides to Every Story: Herpes Simplex Type-1 Viral Glycoproteins gB, gD, gH/gL, gK, and Cellular Receptors Function as Key Players in Membrane Fusion.Viruses. 2021 Sep 16;13(9):1849. doi: 10.3390/v13091849. Viruses. 2021. PMID: 34578430 Free PMC article. Review.
References
-
- King NW, Hunt RD, Daniel MD, Melendez LV. 1967. Overt herpes-T infection in squirrel monkeys (Saimiri sciureus). Lab. Anim. Care 17:413–423 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

