Comparative complete genome analysis of chicken and Turkey megriviruses (family picornaviridae): long 3' untranslated regions with a potential second open reading frame and evidence for possible recombination
- PMID: 24672039
- PMCID: PMC4093843
- DOI: 10.1128/JVI.03807-13
Comparative complete genome analysis of chicken and Turkey megriviruses (family picornaviridae): long 3' untranslated regions with a potential second open reading frame and evidence for possible recombination
Abstract
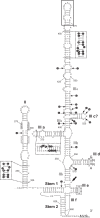
Members of the family Picornaviridae consist of small positive-sense single-stranded RNA (+ssRNA) viruses capable of infecting various vertebrate species, including birds. One of the recently identified avian picornaviruses, with a remarkably long (>9,040-nucleotide) but still incompletely sequenced genome, is turkey hepatitis virus 1 (THV-1; species Melegrivirus A, genus Megrivirus), a virus associated with liver necrosis and enteritis in commercial turkeys (Meleagris gallopavo). This report presents the results of the genetic analysis of three complete genomes of megriviruses from fecal samples of chickens (chicken/B21-CHV/2012/HUN, GenBank accession no. KF961186, and chicken/CHK-IV-CHV/2013/HUN, GenBank accession no. KF961187) (Gallus gallus domesticus) and turkey (turkey/B407-THV/2011/HUN, GenBank accession no. KF961188) (Meleagris gallopavo) with the largest picornavirus genome (up to 9,739 nucleotides) so far described. The close phylogenetic relationship to THV-1 in the nonstructural protein-coding genome region and possession of the same internal ribosomal entry site type (IVB-like) suggest that the study strains belong to the genus Megrivirus. However, the genome comparisons revealed numerous unique variations (e.g., different numbers of potential 2A peptides, unusually long 3' genome parts with various lengths of a potential second open reading frame, and multiple repeating sequence motifs in the 3' untranslated region) and heterogeneous sequence relationships between the structural and nonstructural genome regions. These differences suggest the classification of chicken megrivirus-like viruses into a candidate novel species in the genus Megrivirus. Based on the different phylogenetic positions of chicken megrivirus-like viruses at the structural and nonstructural genome regions, the recombinant nature of these viruses is plausible.
Importance: The comparative genome analysis of turkey and novel chicken megriviruses revealed numerous unique genome features, e.g., up to four potential 2A peptides, unusually long 3' genome parts with various lengths containing a potential second open reading frame, multiple repeating sequence motifs, and heterogeneous sequence relationships (possibly due to a recombination event) between the structural and nonstructural genome regions. Our results could help us to better understand the evolution and diversity (in terms of sequence and genome layout) of picornaviruses.
Figures



References
-
- Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. 2011. Picornaviridae, p 855–880 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom
-
- Racaniello V. 2007. Picornaviridae: the viruses and their replication, p 795–838 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA
-
- Woo PC, Lau SK, Choi GK, Huang Y, Teng JL, Tsoi HW, Tse H, Yeung ML, Chan KH, Jin DY, Yuen KY. 2012. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J. Virol. 86:2797–2808. 10.1128/JVI.05481-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

