An allometric relationship between the genome length and virion volume of viruses
- PMID: 24672040
- PMCID: PMC4093846
- DOI: 10.1128/JVI.00362-14
An allometric relationship between the genome length and virion volume of viruses
Abstract
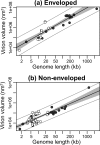
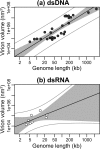
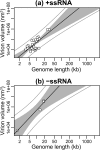
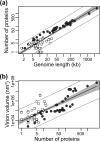
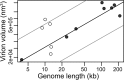
Virions vary in size by at least 4 orders of magnitude, yet the evolutionary forces responsible for this enormous diversity are unknown. We document a significant allometric relationship, with an exponent of approximately 1.5, between the genome length and virion volume of viruses and find that this relationship is not due to geometric constraints. Notably, this allometric relationship holds regardless of genomic nucleic acid, genome structure, or type of virion architecture and therefore represents a powerful scaling law. In contrast, no such relationship is observed at the scale of individual genes. Similarly, after adjusting for genome length, no association is observed between virion volume and the number of proteins, ruling out protein number as the explanation for the relationship between genome and virion sizes. Such a fundamental allometric relationship not only sheds light on the constraints to virus evolution, in that increases in virion size but not necessarily structure are associated with concomitant increases in genome size, but also implies that virion sizes in nature can be broadly predicted from genome sequence data alone.
Importance: Viruses vary dramatically in both genome and virion sizes, but the factors responsible for this diversity are uncertain. Through a comparative and quantitative investigation of these two fundamental biological parameters across diverse viral taxa, we show that genome length and virion volume conform to a simple allometric scaling law. Notably, this allometric relationship holds regardless of the type of virus, including those with both RNA and DNA genomes, and encompasses viruses that exhibit more than 3 logs of genome size variation. Accordingly, this study helps to reveal the basic rules of virus design.
Figures



References
-
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed). 2005. Virus taxonomy; eighth report of the International Committee on Taxonomy of Viruses. Elsevier, London, United Kingdom
-
- Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJ, Schmidt CF. 2006. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc. Natl. Acad. Sci. U. S. A. 103:6184–6189. 10.1073/pnas.0601744103 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

