Vibrio chromosome-specific families
- PMID: 24672511
- PMCID: PMC3957060
- DOI: 10.3389/fmicb.2014.00073
Vibrio chromosome-specific families
Abstract
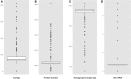
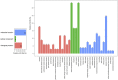
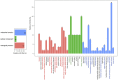
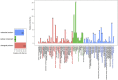
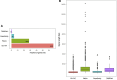
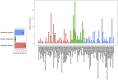
We have compared chromosome-specific genes in a set of 18 finished Vibrio genomes, and, in addition, also calculated the pan- and core-genomes from a data set of more than 250 draft Vibrio genome sequences. These genomes come from 9 known species and 2 unknown species. Within the finished chromosomes, we find a core set of 1269 encoded protein families for chromosome 1, and a core of 252 encoded protein families for chromosome 2. Many of these core proteins are also found in the draft genomes (although which chromosome they are located on is unknown.) Of the chromosome specific core protein families, 1169 and 153 are uniquely found in chromosomes 1 and 2, respectively. Gene ontology (GO) terms for each of the protein families were determined, and the different sets for each chromosome were compared. A total of 363 different "Molecular Function" GO categories were found for chromosome 1 specific protein families, and these include several broad activities: pyridoxine 5' phosphate synthetase, glucosylceramidase, heme transport, DNA ligase, amino acid binding, and ribosomal components; in contrast, chromosome 2 specific protein families have only 66 Molecular Function GO terms and include many membrane-associated activities, such as ion channels, transmembrane transporters, and electron transport chain proteins. Thus, it appears that whilst there are many "housekeeping systems" encoded in chromosome 1, there are far fewer core functions found in chromosome 2. However, the presence of many membrane-associated encoded proteins in chromosome 2 is surprising.
Keywords: Vibrio comparative genomics; Vibrio core-genome; Vibrio pan-genome; chromosome-specific genes; comparative genomics.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

