Prolyl-hydroxylase 3: Evolving Roles for an Ancient Signaling Protein
- PMID: 24672806
- PMCID: PMC3963164
- DOI: 10.2147/HP.S50091
Prolyl-hydroxylase 3: Evolving Roles for an Ancient Signaling Protein
Abstract
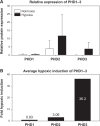
The ability of cells to sense oxygen is a highly evolved process that facilitates adaptations to the local oxygen environment and is critical to energy homeostasis. In vertebrates, this process is largely controlled by three intracellular prolyl-4-hydroxylases (PHD 1-3). These related enzymes share the ability to hydroxylate the hypoxia-inducible transcription factor (HIF), and therefore control the transcription of genes involved in metabolism and vascular recruitment. However, it is becoming increasingly apparent that proline-4-hydroxylation controls much more than HIF signaling, with PHD3 emerging as an exceptionally unique and functionally diverse PHD isoform. In fact, PHD3-mediated hydroxylation has recently been purported to function in such diverse roles as sympathetic neuronal and muscle development, sepsis, glycolytic metabolism, and cell fate. PHD3 expression is also highly distinct from that of the other PHD enzymes, and varies considerably between different cell types and oxygen concentrations. This review will examine the evolution of oxygen sensing by the HIF-family of PHD enzymes, with a specific focus on complex nature of PHD3 expression and function in mammalian cells.
Keywords: EGLN3; HIF-PHD; Hypoxia; Hypoxia Inducible Factor; Oxygen Sensing; PHD3.
Figures








References
-
- Taylor CT, McElwain JC. Ancient atmospheres and the evolution of oxygen sensing via the hypoxia-inducible factor in metazoans. Physiology (Bethesda) 2010;25(5):272–279. - PubMed
-
- Catling DC, Claire MW. How Earth’s atmosphere evolved to an oxic state: a status report. Earth Planet Sci Lett. 2006;237(1–2):1–20.
-
- West CM, van der Wel H, Wang ZA. Prolyl 4-hydroxylase-1 mediates O2 signaling during development of Dictyostelium. Development. 2007;134(18):3349–3358. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

