Genetic variation of the whole ICAM4 gene in Caucasians and African Americans
- PMID: 24673173
- PMCID: PMC4163062
- DOI: 10.1111/trf.12615
Genetic variation of the whole ICAM4 gene in Caucasians and African Americans
Abstract
Background: Landsteiner-Wiener (LW) is the human blood group system Number 16, which comprises two antithetical antigens, LW(a) and LW(b) and the high-prevalence antigen LW(ab) . LW is encoded by the intracellular adhesion molecule 4 (ICAM4) gene. The ICAM4 protein is part of the Rhesus complex in the red cell membrane and is involved in cell-cell adhesion.
Study design and methods: We developed a method to sequence the whole 1.9-kb ICAM4 gene from genomic DNA in one amplicon. We determined the nucleotide sequence of Exons 1 to 3, the two introns, and 402-bp 5'-untranslated region (UTR) and 347-bp 3'-UTR in 97 Caucasian and 91 African American individuals.
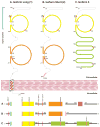
Results: Seven variant ICAM4 alleles were found, distinct from the wild-type ICAM4 allele (GenBank KF712272), known as LW*05 and encoding LW(a) . An effect of the LW(a) /LW(b) amino acid substitution on the protein structure was predicted by two of the three computational modeling programs used.
Conclusions: We describe a practical approach for sequencing and determining the ICAM4 alleles using genomic DNA. LW*05 is the ancestral allele, which had also been observed in a Neanderthal sample. All seven variant alleles are immediate derivatives of the prevalent LW*05 and caused by one single-nucleotide polymorphism (SNP) in each allele. Our data were consistent with the NHLBI GO Exome Sequencing Project (ESP) and the dbSNP databases, as all SNPs had been observed previously. Our study has the advantage over the other databases in that it adds haplotype (allele) information for the ICAM4 gene, clinically relevant in the field of transfusion medicine.
Published 2014. This article is a U.S. Government work and is in the public domain in the USA.
Conflict of interest statement
Figures

References
-
- Storry JR, Castilho L, Daniels G, Flegel WA, Garratty G, de Haas M, Hyland C, Lomas-Francis C, Moulds JM, Nogues N, Olsson ML, Poole J, Reid ME, Rouger P, van der Schoot E, Scott M, Tani Y, Yu LC, Wendel S, Westhoff C, Yahalom V, Zelinski T. International Society of Blood Transfusion Working Party on Red Cell Immunogenetics and Blood Group Terminology: Cancun Report (2012) (in press) Vox Sang. 2013 - PMC - PubMed
-
- Grandstaff Moulds MK. The LW blood group system: a review. Immunohematology. 2011;27:136–42. - PubMed
-
- Byrne KM, Byrne PC. Review: other blood group systems--Diego, Yt, Xg, Scianna, Dombrock, Colton, Landsteiner-Wiener, and Indian. Immunohematology. 2004;20:50–8. - PubMed
-
- Hermand P, Gane P, Mattei MG, Sistonen P, Cartron JP, Bailly P. Molecular basis and expression of the LWa/LWb blood group polymorphism. Blood. 1995;86:1590–4. - PubMed
-
- Gibbs MB. The Quantitative Relationship of the Rh-like (LW) and D Antigens of Human Erythrocytes. Nature. 1966;210:642–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

