High-density nucleosome occupancy map of human chromosome 9p21-22 reveals chromatin organization of the type I interferon gene cluster
- PMID: 24673249
- PMCID: PMC4150395
- DOI: 10.1089/jir.2013.0118
High-density nucleosome occupancy map of human chromosome 9p21-22 reveals chromatin organization of the type I interferon gene cluster
Abstract
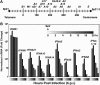
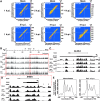
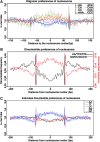
Genome-wide investigations have dramatically increased our understanding of nucleosome positioning and the role of chromatin in gene regulation, yet some genomic regions have been poorly represented in human nucleosome maps. One such region is represented by human chromosome 9p21-22, which contains the type I interferon gene cluster that includes 16 interferon alpha genes and the single interferon beta, interferon epsilon, and interferon omega genes. A high-density nucleosome mapping strategy was used to generate locus-wide maps of the nucleosome organization of this biomedically important locus at a steady state and during a time course of infection with Sendai virus, an inducer of interferon gene expression. Detailed statistical and computational analysis illustrates that nucleosomes in this locus exhibit preferences for particular dinucleotide and oligomer DNA sequence motifs in vivo, which are similar to those reported for lower eukaryotic nucleosome-DNA interactions. These data were used to visualize the region's chromatin architecture and reveal features that are common to the organization of all the type I interferon genes, indicating a common nucleosome-mediated gene regulatory paradigm. Additionally, this study clarifies aspects of the dynamic changes that occur with the nucleosome occupying the transcriptional start site of the interferon beta gene after virus infection.
Figures

References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103(4):667–678 - PubMed
-
- Apostolou E, Thanos D. 2008. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134(1):85–96 - PubMed
-
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD, Ansari AZ, Nislow C, Hughes TR. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32(6):878–887 - PMC - PubMed
-
- Bashiardes S, Veile R, Helms C, Mardis ER, Bowcock AM, Lovett M. 2005. Direct genomic selection. Nat Methods 2(1):63–69 - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

