The ability of thyroid hormone receptors to sense t4 as an agonist depends on receptor isoform and on cellular cofactors
- PMID: 24673558
- PMCID: PMC4004778
- DOI: 10.1210/me.2013-1335
The ability of thyroid hormone receptors to sense t4 as an agonist depends on receptor isoform and on cellular cofactors
Abstract
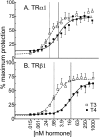
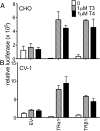
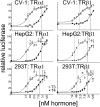
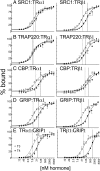
T4 (3,5,3',5'-tetraiodo-l-thyronine) is classically viewed as a prohormone that must be converted to the T3 (3,5,3'-triiodo-l-thyronine) form for biological activity. We first determined that the ability of reporter genes to respond to T4 and to T3 differed for the different thyroid hormone receptor (TR) isoforms, with TRα1 generally more responsive to T4 than was TRβ1. The response to T4 vs T3 also differed dramatically in different cell types in a manner that could not be attributed to differences in deiodinase activity or in hormone affinity, leading us to examine the role of TR coregulators in this phenomenon. Unexpectedly, several coactivators, such as steroid receptor coactivator-1 (SRC1) and thyroid hormone receptor-associated protein 220 (TRAP220), were recruited to TRα1 nearly equally by T4 as by T3 in vitro, indicating that TRα1 possesses an innate potential to respond efficiently to T4 as an agonist. In contrast, release of corepressors, such as the nuclear receptor coreceptor NCoRω, from TRα1 by T4 was relatively inefficient, requiring considerably higher concentrations of this ligand than did coactivator recruitment. Our results suggest that cells, by altering the repertoire and abundance of corepressors and coactivators expressed, may regulate their ability to respond to T4, raising the possibility that T4 may function directly as a hormone in specific cellular or physiological contexts.
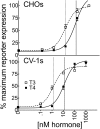
Figures




References
-
- Larsen PR, Davies TF, Schlumberger MJ, Hay ID. Thyroid physiology and diagnostic evaluation of patients with thyroid disorders. In: Larsen PR, Kronenberg HM, Melmed S, Polonsky KS, eds. Williams Textbook of Endocrinology. 10th ed Philadelphia: Saunders; 2003:331–373.
-
- Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097–1142. - PubMed
-
- Flamant F, Baxter JD, Forrest D, et al. International Union of Pharmacology. LIX. The pharmacology and classification of the nuclear receptor superfamily: thyroid hormone receptors. Pharmacol Rev. 2006;58:705–711. - PubMed
-
- Wu Y, Koenig RJ. Gene regulation by thyroid hormone. Trends Endocrinol Metab. 2000;11:207–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

