The ubiquilin gene family: evolutionary patterns and functional insights
- PMID: 24674348
- PMCID: PMC4230246
- DOI: 10.1186/1471-2148-14-63
The ubiquilin gene family: evolutionary patterns and functional insights
Abstract
Background: Ubiquilins are proteins that function as ubiquitin receptors in eukaryotes. Mutations in two ubiquilin-encoding genes have been linked to the genesis of neurodegenerative diseases. However, ubiquilin functions are still poorly understood.
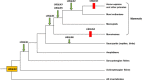
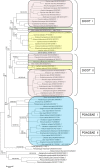
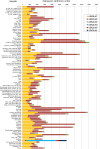
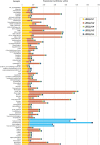
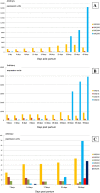
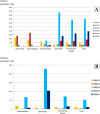
Results: In this study, evolutionary and functional data are combined to determine the origin and diversification of the ubiquilin gene family and to characterize novel potential roles of ubiquilins in mammalian species, including humans. The analysis of more than six hundred sequences allowed characterizing ubiquilin diversity in all the main eukaryotic groups. Many organisms (e. g. fungi, many animals) have single ubiquilin genes, but duplications in animal, plant, alveolate and excavate species are described. Seven different ubiquilins have been detected in vertebrates. Two of them, here called UBQLN5 and UBQLN6, had not been hitherto described. Significantly, marsupial and eutherian mammals have the most complex ubiquilin gene families, composed of up to 6 genes. This exceptional mammalian-specific expansion is the result of the recent emergence of four new genes, three of them (UBQLN3, UBQLN5 and UBQLNL) with precise testis-specific expression patterns that indicate roles in the postmeiotic stages of spermatogenesis. A gene with related features has independently arisen in species of the Drosophila genus. Positive selection acting on some mammalian ubiquilins has been detected.
Conclusions: The ubiquilin gene family is highly conserved in eukaryotes. The infrequent lineage-specific amplifications observed may be linked to the emergence of novel functions in particular tissues.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

