Comparative proteomic analysis of human lung telocytes with fibroblasts
- PMID: 24674459
- PMCID: PMC4000110
- DOI: 10.1111/jcmm.12290
Comparative proteomic analysis of human lung telocytes with fibroblasts
Abstract
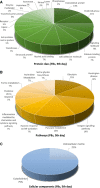
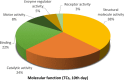
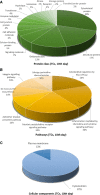
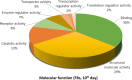
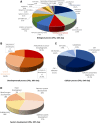
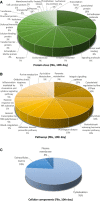
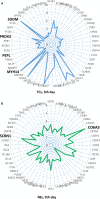
Telocytes (TCs) were recently described as interstitial cells with very long prolongations named telopodes (Tps; www.telocytes.com). Establishing the TC proteome is a priority to show that TCs are a distinct type of cells. Therefore, we examined the molecular aspects of lung TCs by comparison with fibroblasts (FBs). Proteins extracted from primary cultures of these cells were analysed by automated 2-dimensional nano-electrospray ionization liquid chromatography tandem mass spectrometry (2D Nano-ESI LC-MS/MS). Differentially expressed proteins were screened by two-sample t-test (P < 0.05) and fold change (>2), based on the bioinformatics analysis. We identified hundreds of proteins up- or down-regulated, respectively, in TCs as compared with FBs. TC proteins with known identities are localized in the cytoskeleton (87%) and plasma membrane (13%), while FB up-regulated proteins are in the cytoskeleton (75%) and destined to extracellular matrix (25%). These identified proteins were classified into different categories based on their molecular functions and biological processes. While the proteins identified in TCs are mainly involved in catalytic activity (43%) and as structural molecular activity (25%), the proteins in FBs are involved in catalytic activity (24%) and in structural molecular activity, particularly synthesis of collagen and other extracellular matrix components (25%). Anyway, our data show that TCs are completely different from FBs. In conclusion, we report here the first extensive identification of proteins from TCs using a quantitative proteomics approach. Protein expression profile shows many up-regulated proteins e.g. myosin-14, periplakin, suggesting that TCs might play specific roles in mechanical sensing and mechanochemical conversion task, tissue homoeostasis and remodelling/renewal. Furthermore, up-regulated proteins matching those found in extracellular vesicles emphasize TCs roles in intercellular signalling and stem cell niche modulation. The novel proteins identified in TCs will be an important resource for further proteomic research and it will possibly allow biomarker identification for TCs. It also creates the premises for understanding the pathogenesis of some lung diseases involving TCs.
Keywords: LC-MS/MS; fibroblasts; isobaric tags for relative and absolute quantification (iTRAQ); lung; proteomics; telocytes.
© 2014 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures









References
-
- Suciu L, Popescu LM, Gherghiceanu M, et al. Telocytes in human term placenta: morphology and phenotype. Cells Tissues Organs. 2010;192:325–39. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

