Total synthesis of a functional designer eukaryotic chromosome
- PMID: 24674868
- PMCID: PMC4033833
- DOI: 10.1126/science.1249252
Total synthesis of a functional designer eukaryotic chromosome
Erratum in
- Science. 2014 May 23;344(6186):816
Abstract
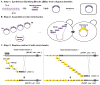
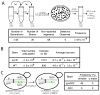
Rapid advances in DNA synthesis techniques have made it possible to engineer viruses, biochemical pathways and assemble bacterial genomes. Here, we report the synthesis of a functional 272,871-base pair designer eukaryotic chromosome, synIII, which is based on the 316,617-base pair native Saccharomyces cerevisiae chromosome III. Changes to synIII include TAG/TAA stop-codon replacements, deletion of subtelomeric regions, introns, transfer RNAs, transposons, and silent mating loci as well as insertion of loxPsym sites to enable genome scrambling. SynIII is functional in S. cerevisiae. Scrambling of the chromosome in a heterozygous diploid reveals a large increase in a-mater derivatives resulting from loss of the MATα allele on synIII. The complete design and synthesis of synIII establishes S. cerevisiae as the basis for designer eukaryotic genome biology.
Conflict of interest statement
The authors declare no competing financial interests.
“This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at
Figures


Comment in
-
Synthetic biology: Construction of a yeast chromosome.Nature. 2014 May 8;509(7499):168-9. doi: 10.1038/509168a. Nature. 2014. PMID: 24805340 No abstract available.
-
Synthetic yeast chromosomes help probe mysteries of evolution.Nature. 2017 Mar 9;543(7645):298-299. doi: 10.1038/nature.2017.21615. Nature. 2017. PMID: 28300123 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

