An immunomics approach to schistosome antigen discovery: antibody signatures of naturally resistant and chronically infected individuals from endemic areas
- PMID: 24675823
- PMCID: PMC3968167
- DOI: 10.1371/journal.ppat.1004033
An immunomics approach to schistosome antigen discovery: antibody signatures of naturally resistant and chronically infected individuals from endemic areas
Abstract
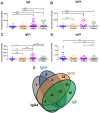
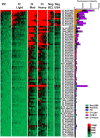
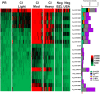
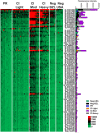
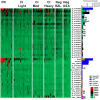
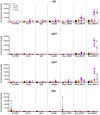
Schistosomiasis is a neglected tropical disease that is responsible for almost 300,000 deaths annually. Mass drug administration (MDA) is used worldwide for the control of schistosomiasis, but chemotherapy fails to prevent reinfection with schistosomes, so MDA alone is not sufficient to eliminate the disease, and a prophylactic vaccine is required. Herein, we take advantage of recent advances in systems biology and longitudinal studies in schistosomiasis endemic areas in Brazil to pilot an immunomics approach to the discovery of schistosomiasis vaccine antigens. We selected mostly surface-derived proteins, produced them using an in vitro rapid translation system and then printed them to generate the first protein microarray for a multi-cellular pathogen. Using well-established Brazilian cohorts of putatively resistant (PR) and chronically infected (CI) individuals stratified by the intensity of their S. mansoni infection, we probed arrays for IgG subclass and IgE responses to these antigens to detect antibody signatures that were reflective of protective vs. non-protective immune responses. Moreover, probing for IgE responses allowed us to identify antigens that might induce potentially deleterious hypersensitivity responses if used as subunit vaccines in endemic populations. Using multi-dimensional cluster analysis we showed that PR individuals mounted a distinct and robust IgG1 response to a small set of newly discovered and well-characterized surface (tegument) antigens in contrast to CI individuals who mounted strong IgE and IgG4 responses to many antigens. Herein, we show the utility of a vaccinomics approach that profiles antibody responses of resistant individuals in a high-throughput multiplex approach for the identification of several potentially protective and safe schistosomiasis vaccine antigens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- van der Werf MJ, de Vlas SJ, Brooker S, Looman CWN, Nagelkerke NJD, et al. (2003) Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop 86: 125–139. - PubMed
-
- King CH, Dickman K, Tisch DJ (2005) Reassessment of the cost of chronic helmintic infection: a meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet 365: 1561–1569. - PubMed
-
- King CH, Dangerfield-Cha M (2008) The unacknowledged impact of chronic schistosomiasis. Chronic Illn 4: 65–79. - PubMed
-
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J (2006) Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis 6: 411–425. - PubMed
-
- Gray DJ, McManus DP, Li Y, Williams GM, Bergquist R, et al. (2010) Schistosomiasis elimination: lessons from the past guide the future. Lancet Infect Dis 10: 733–736. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

