Microfluidic source-sink model reveals effects of biophysically distinct CXCL12 isoforms in breast cancer chemotaxis
- PMID: 24675873
- PMCID: PMC4034763
- DOI: 10.1039/c4ib00015c
Microfluidic source-sink model reveals effects of biophysically distinct CXCL12 isoforms in breast cancer chemotaxis
Abstract
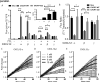
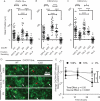
Chemokines critically regulate chemotaxis in normal and pathologic states, but there is limited understanding of how multicellular interactions generate gradients needed for cell migration. Previous studies of chemotaxis of CXCR4+ cells toward chemokine CXCL12 suggest the requirement of cells expressing scavenger receptor CXCR7 in a source-sink system. We leveraged an established microfluidic device to discover that chemotaxis of CXCR4 cells toward distinct isoforms of CXCL12 required CXCR7 scavenging only under conditions with higher than optimal levels of CXCL12. Chemotaxis toward CXCL12-β and -γ isoforms, which have greater binding to extracellular molecules and have been largely overlooked, was less dependent on CXCR7 than the more commonly studied CXCL12-α. Chemotaxis of CXCR4+ cells toward even low levels of CXCL12-γ and CXCL12-β still occurred during treatment with a FDA-approved inhibitor of CXCR4. We also detected CXCL12-γ only in breast cancers from patients with advanced disease. Physiological gradient formation within the device facilitated interrogation of key differences in chemotaxis among CXCL12 isoforms and suggests CXCL12-γ as a biomarker for metastatic cancer.
Figures


References
-
- Raman D, Sobolik-Delmaire T, Richmond A. Chemokines in health and disease. Experimental Cell Research. 2011;317(5):575–589. - PMC - PubMed
- Burkhardt AM, Homey B, Zlotnik A. Homeostatic chemokine receptors and organ-specific metastasis. Nature Reviews Immunology. 2011;11(9):597+. - PubMed
- Balkwill FR. The chemokine system and cancer. The Journal of Pathology. 2012;226(2):148–157. - PubMed
- Zhang GX, Baker CM, Kolson DL, Rostami AM. Chemokines and chemokine receptors in the pathogenesis of multiple sclerosis. Multiple Sclerosis. 2000;6(1):3–13. - PubMed
- Zernecke A, Shagdarsuren E, Weber C. Chemokines in atherosclerosis: an update. Arterioscler Throm Vasc Biol. 2008;28(11):1897–1908. - PubMed
- Krumbholz M, Theil D, Cepok S, Hemmer B, Kivisäkk P, Ransohoff RM, Hofbauer M, Farina C, Derfuss T, Hartle C, Newcombe J, Hohlfeld R, Meinl E. Chemokines in multiple sclerosis: CXCL12 and CXCL13 up-regulation is differentially linked to CNS immune cell recruitment. Brain. 2006;129(1):200–211. - PubMed
-
- Yu SR, Burkhardt M, Nowak M, Ries J, Petrasek Z, Scholpp S, Schwille P, Brand M. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461(7263):533–6. doi: 10.1038/nature08391. Epub 2009 Sep 9. - PubMed
- Boldajipour B, Mahabaleshwar S, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132(3):463–473. - PubMed
- Torisawa Y, Mosadegh B, Bersano-Begey T, Steele J, Luker K, Luker G, Takayama S. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Integr Biol (Camb) 2010;2(11-12):680–686. - PMC - PubMed
-
- Scholpp S, Brand M. Endocytosis controls spreading and effective signaling range of Fgf8 protein. Current biology. 2004;14(20):1834–1841. - PubMed
- Naumann U, Cameroni E, Pruenster M, Mahabaleshwar S, Raz E, Zerwes H, Rot A, Thelen M. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5(2):e9175. - PMC - PubMed
- Luker K, Steele J, Mihalko L, Luker G. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599–4610. - PMC - PubMed
- Luker K, Lewin S, Mihalko L, Schmidt B, Winkler J, Coggins N, Thomas D, Luker G. Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene. 2012 doi: 10.1038/onc.2011.633. [Epub ahead of print] - PMC - PubMed
- Ray P, Mihalko L, Coggins N, Moudgil P, Ehrlich A, Luker K, Luker G. Carboxy-terminus of CXCR7 regulates receptor localization and function. Int J Biochem Cell Biol. 2012;44(4):669–678. - PMC - PubMed
- Fredericks Z, Pitcher J, Lefkowitz R. Identification of the G protein-coupled receptor kinase phosphorylation sites in the human beta2-adrenergic receptor. J Biol Chem. 1996;271(23):13796–13803. - PubMed
- Sanchez-Alcaniz J, Haege S, Mueller W, Pla R, Mackay F, Schulz S, Lopez-Bendito G, Stumm R, Marin O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69(1):77–90. - PubMed
- Wang Y, Li G, Stanco A, Long J, Crawford D, Potter G, Pleasure S, Behrens T, Rubenstein J. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69(1):61–76. - PMC - PubMed
-
- Weber M, Hauschild R, Schwarz J, Moussion C, de Vries I, Legler DF, Luther SA, Bollenbach T, Sixt M. Interstitial Dendritic Cell Guidance by Haptotactic Chemokine Gradients. Science. 2013;339(6117):328–332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR028819/RR/NCRR NIH HHS/United States
- R01CA136553/CA/NCI NIH HHS/United States
- R01CA136829/CA/NCI NIH HHS/United States
- R01 CA136829/CA/NCI NIH HHS/United States
- R01EB012579/EB/NIBIB NIH HHS/United States
- R01 CA170198/CA/NCI NIH HHS/United States
- R01CA142750/CA/NCI NIH HHS/United States
- R01 CA142750/CA/NCI NIH HHS/United States
- R01 CA136553/CA/NCI NIH HHS/United States
- P50CA093990/CA/NCI NIH HHS/United States
- T32 CA140044/CA/NCI NIH HHS/United States
- P50 CA093990/CA/NCI NIH HHS/United States
- R01GM096040/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

