Structure of the yeast mitochondrial large ribosomal subunit
- PMID: 24675956
- PMCID: PMC4046073
- DOI: 10.1126/science.1249410
Structure of the yeast mitochondrial large ribosomal subunit
Abstract
Mitochondria have specialized ribosomes that have diverged from their bacterial and cytoplasmic counterparts. We have solved the structure of the yeast mitoribosomal large subunit using single-particle cryo-electron microscopy. The resolution of 3.2 angstroms enabled a nearly complete atomic model to be built de novo and refined, including 39 proteins, 13 of which are unique to mitochondria, as well as expansion segments of mitoribosomal RNA. The structure reveals a new exit tunnel path and architecture, unique elements of the E site, and a putative membrane docking site.
Figures

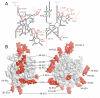



Comment in
-
Biochemistry. The resolution revolution.Science. 2014 Mar 28;343(6178):1443-4. doi: 10.1126/science.1251652. Science. 2014. PMID: 24675944 No abstract available.
References
-
- Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2010;1803:767. - PubMed
-
- Jones CN, Miller C, Tenenbaum A, Spremulli LL, Saada A. Antibiotic effects on mitochondrial translation and in patients with mitochondrial translational defects. Mitochondrion. 2009;9:429. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

