Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing
- PMID: 24676216
- PMCID: PMC3967949
- DOI: 10.1371/journal.pgen.1004271
Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing
Abstract
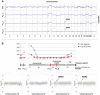
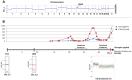
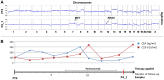
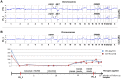
Monoclonal antibodies targeting the Epidermal Growth Factor Receptor (EGFR), such as cetuximab and panitumumab, have evolved to important therapeutic options in metastatic colorectal cancer (CRC). However, almost all patients with clinical response to anti-EGFR therapies show disease progression within a few months and little is known about mechanism and timing of resistance evolution. Here we analyzed plasma DNA from ten patients treated with anti-EGFR therapy by whole genome sequencing (plasma-Seq) and ultra-sensitive deep sequencing of genes associated with resistance to anti-EGFR treatment such as KRAS, BRAF, PIK3CA, and EGFR. Surprisingly, we observed that the development of resistance to anti-EGFR therapies was associated with acquired gains of KRAS in four patients (40%), which occurred either as novel focal amplifications (n = 3) or as high level polysomy of 12p (n = 1). In addition, we observed focal amplifications of other genes recently shown to be involved in acquired resistance to anti-EGFR therapies, such as MET (n = 2) and ERBB2 (n = 1). Overrepresentation of the EGFR gene was associated with a good initial anti-EGFR efficacy. Overall, we identified predictive biomarkers associated with anti-EGFR efficacy in seven patients (70%), which correlated well with treatment response. In contrast, ultra-sensitive deep sequencing of KRAS, BRAF, PIK3CA, and EGFR did not reveal the occurrence of novel, acquired mutations. Thus, plasma-Seq enables the identification of novel mutant clones and may therefore facilitate early adjustments of therapies that may delay or prevent disease progression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, et al. (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765. - PubMed
-
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, et al. (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360: 1408–1417. - PubMed
-
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, et al. (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634. - PubMed
-
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, et al. (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 351: 337–345. - PubMed
-
- Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, et al. (2007) Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 25: 1658–1664. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

