Revising the Representation of Fatty Acid, Glycerolipid, and Glycerophospholipid Metabolism in the Consensus Model of Yeast Metabolism
- PMID: 24678285
- PMCID: PMC3963290
- DOI: 10.1089/ind.2013.0013
Revising the Representation of Fatty Acid, Glycerolipid, and Glycerophospholipid Metabolism in the Consensus Model of Yeast Metabolism
Abstract
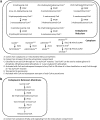
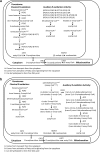
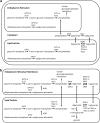
Genome-scale metabolic models are built using information from an organism's annotated genome and, correspondingly, information on reactions catalyzed by the set of metabolic enzymes encoded by the genome. These models have been successfully applied to guide metabolic engineering to increase production of metabolites of industrial interest. Congruity between simulated and experimental metabolic behavior is influenced by the accuracy of the representation of the metabolic network in the model. In the interest of applying the consensus model of Saccharomyces cerevisiae metabolism for increased productivity of triglycerides, we manually evaluated the representation of fatty acid, glycerophospholipid, and glycerolipid metabolism in the consensus model (Yeast v6.0). These areas of metabolism were chosen due to their tightly interconnected nature to triglyceride synthesis. Manual curation was facilitated by custom MATLAB functions that return information contained in the model for reactions associated with genes and metabolites within the stated areas of metabolism. Through manual curation, we have identified inconsistencies between information contained in the model and literature knowledge. These inconsistencies include incorrect gene-reaction associations, improper definition of substrates/products in reactions, inappropriate assignments of reaction directionality, nonfunctional β-oxidation pathways, and missing reactions relevant to the synthesis and degradation of triglycerides. Suggestions to amend these inconsistencies in the Yeast v6.0 model can be implemented through a MATLAB script provided in theSupplementary Materials, Supplementary Data S1(Supplementary Data are available online at www.liebertpub.com/ind).
Figures

References
-
- US Environmental Protection Agency. Biofuels and the environment: First triennial report to Congress; 2011. EPA/600/R-10/183F.
-
- Kosa M. Ragauskas AJ. Lipids from heterotrophic microbes: Advances in metabolism research. Trends Biotechnol. 2011;29(2):53–61. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
