Evolving models for peroxisome biogenesis
- PMID: 24681485
- PMCID: PMC4148619
- DOI: 10.1016/j.ceb.2014.02.002
Evolving models for peroxisome biogenesis
Abstract
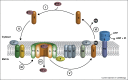
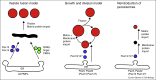
Significant progress has been made towards our understanding of the mechanism of peroxisome formation, in particular concerning sorting of peroxisomal membrane proteins, matrix protein import and organelle multiplication. Here we evaluate the progress made in recent years. We focus mainly on progress made in yeasts. We indicate the gaps in our knowledge and discuss conflicting models.
Crown Copyright © 2014. Published by Elsevier Ltd. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

