A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase
- PMID: 24681617
- PMCID: PMC4001383
- DOI: 10.1105/tpc.114.124495
A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase
Abstract
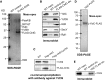
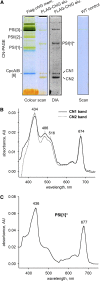
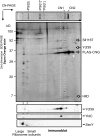
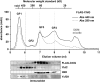
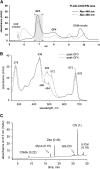
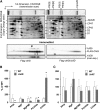
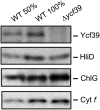
Macromolecular membrane assemblies of chlorophyll-protein complexes efficiently harvest and trap light energy for photosynthesis. To investigate the delivery of chlorophylls to the newly synthesized photosystem apoproteins, a terminal enzyme of chlorophyll biosynthesis, chlorophyll synthase (ChlG), was tagged in the cyanobacterium Synechocystis PCC 6803 (Synechocystis) and used as bait in pull-down experiments. We retrieved an enzymatically active complex comprising ChlG and the high-light-inducible protein HliD, which associates with the Ycf39 protein, a putative assembly factor for photosystem II, and with the YidC/Alb3 insertase. 2D electrophoresis and immunoblotting also provided evidence for the presence of SecY and ribosome subunits. The isolated complex contained chlorophyll, chlorophyllide, and carotenoid pigments. Deletion of hliD elevated the level of the ChlG substrate, chlorophyllide, more than 6-fold; HliD is apparently required for assembly of FLAG-ChlG into larger complexes with other proteins such as Ycf39. These data reveal a link between chlorophyll biosynthesis and the Sec/YidC-dependent cotranslational insertion of nascent photosystem polypeptides into membranes. We expect that this close physical linkage coordinates the arrival of pigments and nascent apoproteins to produce photosynthetic pigment-protein complexes with minimal risk of accumulating phototoxic unbound chlorophylls.
Figures

Similar articles
-
Xanthophyll carotenoids stabilise the association of cyanobacterial chlorophyll synthase with the LHC-like protein HliD.Biochem J. 2020 Oct 30;477(20):4021-4036. doi: 10.1042/BCJ20200561. Biochem J. 2020. PMID: 32990304
-
Plant and algal chlorophyll synthases function in Synechocystis and interact with the YidC/Alb3 membrane insertase.FEBS Lett. 2018 Sep;592(18):3062-3073. doi: 10.1002/1873-3468.13222. Epub 2018 Sep 6. FEBS Lett. 2018. PMID: 30107031 Free PMC article.
-
High-light-inducible proteins control associations between chlorophyll synthase and the Photosystem II biogenesis factor Ycf39.Plant Physiol. 2025 May 30;198(2):kiaf213. doi: 10.1093/plphys/kiaf213. Plant Physiol. 2025. PMID: 40413779 Free PMC article.
-
The terminal enzymes of (bacterio)chlorophyll biosynthesis.R Soc Open Sci. 2022 May 4;9(5):211903. doi: 10.1098/rsos.211903. eCollection 2022 May. R Soc Open Sci. 2022. PMID: 35573041 Free PMC article. Review.
-
Making proteins green; biosynthesis of chlorophyll-binding proteins in cyanobacteria.Photosynth Res. 2014 Feb;119(1-2):223-32. doi: 10.1007/s11120-013-9797-2. Epub 2013 Feb 4. Photosynth Res. 2014. PMID: 23377990 Review.
Cited by
-
Subcellular Localization of Carotenoid Biosynthesis in Synechocystis sp. PCC 6803.PLoS One. 2015 Jun 17;10(6):e0130904. doi: 10.1371/journal.pone.0130904. eCollection 2015. PLoS One. 2015. PMID: 26083372 Free PMC article.
-
Advances in the Understanding of the Lifecycle of Photosystem II.Microorganisms. 2022 Apr 19;10(5):836. doi: 10.3390/microorganisms10050836. Microorganisms. 2022. PMID: 35630282 Free PMC article. Review.
-
The Ribosome-Bound Protein Pam68 Promotes Insertion of Chlorophyll into the CP47 Subunit of Photosystem II.Plant Physiol. 2018 Apr;176(4):2931-2942. doi: 10.1104/pp.18.00061. Epub 2018 Feb 20. Plant Physiol. 2018. PMID: 29463774 Free PMC article.
-
Stable Accumulation of Photosystem II Requires ONE-HELIX PROTEIN1 (OHP1) of the Light Harvesting-Like Family.Plant Physiol. 2018 Mar;176(3):2277-2291. doi: 10.1104/pp.17.01782. Epub 2018 Feb 1. Plant Physiol. 2018. PMID: 29438089 Free PMC article.
-
Identification and Roles of Photosystem II Assembly, Stability, and Repair Factors in Arabidopsis.Front Plant Sci. 2016 Feb 16;7:168. doi: 10.3389/fpls.2016.00168. eCollection 2016. Front Plant Sci. 2016. PMID: 26909098 Free PMC article. Review.
References
-
- Antonoaea R., Fürst M., Nishiyama K., Müller M. (2008). The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47: 5649–5656. - PubMed
-
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. - PubMed
-
- Bhaya D., Dufresne A., Vaulot D., Grossman A. (2002). Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiol. Lett. 215: 209–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

