Characterization and evolution of tetrameric photosystem I from the thermophilic cyanobacterium Chroococcidiopsis sp TS-821
- PMID: 24681621
- PMCID: PMC4001380
- DOI: 10.1105/tpc.113.120782
Characterization and evolution of tetrameric photosystem I from the thermophilic cyanobacterium Chroococcidiopsis sp TS-821
Abstract
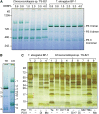
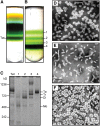
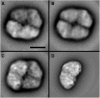
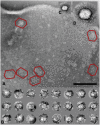
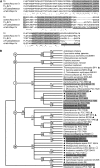
Photosystem I (PSI) is a reaction center associated with oxygenic photosynthesis. Unlike the monomeric reaction centers in green and purple bacteria, PSI forms trimeric complexes in most cyanobacteria with a 3-fold rotational symmetry that is primarily stabilized via adjacent PsaL subunits; however, in plants/algae, PSI is monomeric. In this study, we discovered a tetrameric form of PSI in the thermophilic cyanobacterium Chroococcidiopsis sp TS-821 (TS-821). In TS-821, PSI forms tetrameric and dimeric species. We investigated these species by Blue Native PAGE, Suc density gradient centrifugation, 77K fluorescence, circular dichroism, and single-particle analysis. Transmission electron microscopy analysis of native membranes confirms the presence of the tetrameric PSI structure prior to detergent solubilization. To investigate why TS-821 forms tetramers instead of trimers, we cloned and analyzed its psaL gene. Interestingly, this gene product contains a short insert between the second and third predicted transmembrane helices. Phylogenetic analysis based on PsaL protein sequences shows that TS-821 is closely related to heterocyst-forming cyanobacteria, some of which also have a tetrameric form of PSI. These results are discussed in light of chloroplast evolution, and we propose that PSI evolved stepwise from a trimeric form to tetrameric oligomer en route to becoming monomeric in plants/algae.
Figures
Similar articles
-
Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.Plant Commun. 2021 Oct 13;3(1):100248. doi: 10.1016/j.xplc.2021.100248. eCollection 2022 Jan 10. Plant Commun. 2021. PMID: 35059628 Free PMC article.
-
Comparisons of Electron Transfer Reactions in a Cyanobacterial Tetrameric and Trimeric Photosystem I Complexes.Photochem Photobiol. 2018 May;94(3):564-569. doi: 10.1111/php.12886. Epub 2018 Mar 2. Photochem Photobiol. 2018. PMID: 29315587
-
Physiological and evolutionary implications of tetrameric photosystem I in cyanobacteria.Nat Plants. 2019 Dec;5(12):1309-1319. doi: 10.1038/s41477-019-0566-x. Epub 2019 Dec 9. Nat Plants. 2019. PMID: 31819227
-
Long-wavelength chlorophylls in photosystem I of cyanobacteria: origin, localization, and functions.Biochemistry (Mosc). 2014 Mar;79(3):213-20. doi: 10.1134/S0006297914030067. Biochemistry (Mosc). 2014. PMID: 24821447 Review.
-
Structure and functional role of supercomplexes of IsiA and Photosystem I in cyanobacterial photosynthesis.FEBS Lett. 2005 Jun 13;579(15):3253-7. doi: 10.1016/j.febslet.2005.03.051. Epub 2005 Apr 7. FEBS Lett. 2005. PMID: 15943969 Review.
Cited by
-
Changes in supramolecular organization of cyanobacterial thylakoid membrane complexes in response to far-red light photoacclimation.Sci Adv. 2022 Feb 11;8(6):eabj4437. doi: 10.1126/sciadv.abj4437. Epub 2022 Feb 9. Sci Adv. 2022. PMID: 35138895 Free PMC article.
-
Crystal Structure of Photosystem I Monomer From Synechocystis PCC 6803.Front Plant Sci. 2019 Jan 4;9:1865. doi: 10.3389/fpls.2018.01865. eCollection 2018. Front Plant Sci. 2019. PMID: 30662446 Free PMC article.
-
Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy.Nat Commun. 2019 Oct 30;10(1):4929. doi: 10.1038/s41467-019-12942-8. Nat Commun. 2019. PMID: 31666526 Free PMC article.
-
Photosystem I oligomerization affects lipid composition in Synechocystis sp. PCC 6803.Biochim Biophys Acta Mol Cell Biol Lipids. 2019 Oct;1864(10):1384-1395. doi: 10.1016/j.bbalip.2019.06.013. Epub 2019 Jun 20. Biochim Biophys Acta Mol Cell Biol Lipids. 2019. PMID: 31228574 Free PMC article.
-
Photosystem I: A Paradigm for Understanding Biological Environmental Adaptation Mechanisms in Cyanobacteria and Algae.Int J Mol Sci. 2024 Aug 12;25(16):8767. doi: 10.3390/ijms25168767. Int J Mol Sci. 2024. PMID: 39201454 Free PMC article. Review.
References
-
- Amunts A., Nelson N. (2008). Functional organization of a plant Photosystem I: Evolution of a highly efficient photochemical machine. Plant Physiol. Biochem. 46: 228–237. - PubMed
-
- Amunts A., Nelson N. (2009). Plant photosystem I design in the light of evolution. Structure 17: 637–650. - PubMed
-
- Aspinwall C.L., Sarcina M., Mullineaux C.W. (2004). Phycobilisome mobility in the cyanobacterium Synechococcus sp. PCC7942 is influenced by the trimerisation of Photosystem I. Photosynth. Res. 79: 179–187. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

