SLC25A1, or CIC, is a novel transcriptional target of mutant p53 and a negative tumor prognostic marker
- PMID: 24681808
- PMCID: PMC4012738
- DOI: 10.18632/oncotarget.1831
SLC25A1, or CIC, is a novel transcriptional target of mutant p53 and a negative tumor prognostic marker
Abstract
Mutations of the p53 gene hallmark many human cancers. Several p53 mutant proteins acquire the capability to promote cancer progression and metastasis, a phenomenon defined as Gain of Oncogenic Function (GOF). The downstream targets by which GOF p53 mutants perturb cellular programs relevant to oncogenesis are only partially known. We have previously demonstrated that SLC25A1 (CIC) promotes tumorigenesis, while its inhibition blunts tumor growth. We now report that CIC is a direct transcriptional target of several p53 mutants. We identify a novel interaction between mutant p53 (mutp53) and the transcription factor FOXO-1 which is responsible for regulation of CIC expression levels. Tumor cells harboring mutp53 display higher CIC levels relative to p53 null or wild-type tumors, and inhibition of CIC activity blunts mutp53-driven tumor growth, partially overcoming GOF activity. CIC inhibition also enhances the chemotherapeutic potential of platinum-based agents. Finally, we found that elevated CIC levels predict poor survival outcome in tumors hallmarked by high frequency of p53 mutations. Our results identify CIC as a novel target of mutp53 and imply that the employment of CIC inhibitors may improve survival rates and reduce chemo-resistance in tumors harboring these types of mutations, which are among the most intractable forms of cancers.
Figures


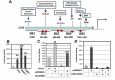



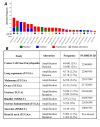
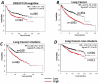
References
-
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. - PubMed
-
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. - PubMed
-
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

