The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells
- PMID: 24682267
- PMCID: PMC4014841
- DOI: 10.1182/blood-2013-07-517177
The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells
Abstract
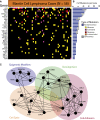
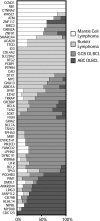
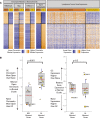
In this study, we define the genetic landscape of mantle cell lymphoma (MCL) through exome sequencing of 56 cases of MCL. We identified recurrent mutations in ATM, CCND1, MLL2, and TP53. We further identified a number of novel genes recurrently mutated in patients with MCL including RB1, WHSC1, POT1, and SMARCA4. We noted that MCLs have a distinct mutational profile compared with lymphomas from other B-cell stages. The ENCODE project has defined the chromatin structure of many cell types. However, a similar characterization of primary human mature B cells has been lacking. We defined, for the first time, the chromatin structure of primary human naïve, germinal center, and memory B cells through chromatin immunoprecipitation and sequencing for H3K4me1, H3K4me3, H3Ac, H3K36me3, H3K27me3, and PolII. We found that somatic mutations that occur more frequently in either MCLs or Burkitt lymphomas were associated with open chromatin in their respective B cells of origin, naïve B cells, and germinal center B cells. Our work thus elucidates the landscape of gene-coding mutations in MCL and the critical interplay between epigenetic alterations associated with B-cell differentiation and the acquisition of somatic mutations in cancer.
Figures

References
-
- Swerdlow SH, Campo E, Harris NL, et al. 4th ed. Lyon, France: IARC Press; 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues.
-
- Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–1971. - PubMed
-
- Meissner B, Kridel R, Lim RS, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood. 2013;121(16):3161–3164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

