Molecular characterization and evolution of a gene family encoding both female- and male-specific reproductive proteins in Drosophila
- PMID: 24682282
- PMCID: PMC4032134
- DOI: 10.1093/molbev/msu114
Molecular characterization and evolution of a gene family encoding both female- and male-specific reproductive proteins in Drosophila
Abstract
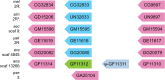
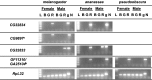
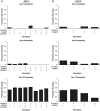
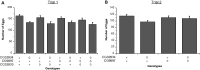
Gene duplication is an important mechanism for the evolution of new reproductive proteins. However, in most cases, each resulting paralog continues to function within the same sex. To investigate the possibility that seminal fluid proteins arise through duplicates of female reproductive genes that become "co-opted" by males, we screened female reproductive genes in Drosophila melanogaster for cases of duplication in which one of the resulting paralogs produces a protein in males that is transferred to females during mating. We identified a set of three tandemly duplicated genes that encode secreted serine-type endopeptidase homologs, two of which are expressed primarily in the female reproductive tract (RT), whereas the third is expressed specifically in the male RT and encodes a seminal fluid protein. Evolutionary and gene expression analyses across Drosophila species suggest that this family arose from a single-copy gene that was female-specific; after duplication, one paralog evolved male-specific expression. Functional tests of knockdowns of each gene in D. melanogaster show that one female-expressed gene is essential for full fecundity, and both female-expressed genes contribute singly or in combination to a female's propensity to remate. In contrast, knockdown of the male-expressed paralog had no significant effect on female fecundity or remating. These data are consistent with a model in which members of this gene family exert effects on females by acting on a common, female-expressed target. After duplication and male co-option of one paralog, the evolution of the interacting proteins could have resulted in differential strengths or effects of each paralog.
Keywords: Drosophila; gene duplication; protease; seminal proteins; sex-specific expression; spermathecal proteins.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Allen AK, Spradling AC. The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development. 2008;135:311–321. - PubMed
-
- Arbeitman MN, Fleming AA, Siegal ML, Null BH, Baker BS. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development. 2004;131:2007–2021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

