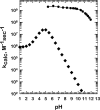
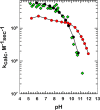
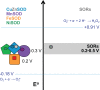
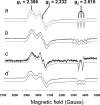
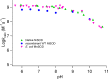
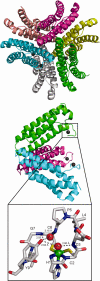
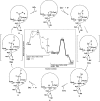
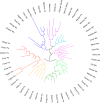
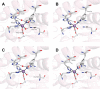
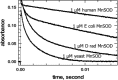
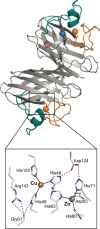
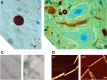
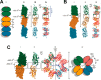
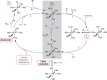
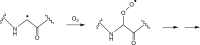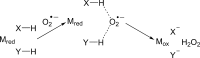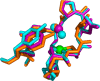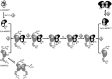Figure 18
Unrooted dendogram of 53 members of the
FeSOD and MnSOD family
wherein branches are colored as follows (clockwise from top left):
blue for mitochondrial MnSODs, magenta for archaeal SODs, teal for
actinobacterial SODs, pink for bacterial MnSODs, light green for cyanobacterial
FeSODs, dark green for FeSODs of plants and green algae, red for FeSODs
of protists, and orange for FeSODs of bacteria. Sequences were chosen
to represent diverse groups of organisms and different metal specificities. BLAST searches of the nonredundant database
of the National Center for Biotechnology and Information (NCBI) were
used to identify additional SOD sequences from weakly represented
groups, and, in those cases in which sequences were very similar,
only one exemplar was retained, the one for which the best information
on metal ion use was available. Where possible, for bacterial and
archaeal SODs especially, the identity of an SOD as Fe-dependent versus
Mn-dependent was sought in primary literature, and the means by which
its metal ion identity was determined is listed as “Anal”
for direct analysis via atomic absorption or another spectroscopic
method, or “H2O2” when it was
inferred on the basis of the SOD’s sensitivity or resistance
to inactivation by H2O2 and a reference is provided.
Some Fe/MnSODs are included, but given that the motivation of this
exercise was to identify residues that correlate differentially with
Fe or Mn use, others are described via Table 4 instead. The tree was displayed and colored using the interactive
tree of life server hosted by the European Molecular Biology Laboratory. The multiple sequence alignment upon which
it is based was generated using MUSCLE (in the “full” most stringent mode) for up to 16 interactions,
as accessed via Phylogeny.fr hosted by the Centre National de la Recherche
Scientifique. The alignment was curated
using Gblocks at the most stringent
setting (not allowing many contiguous nonconserved positions), and
the results were inspected visually via the Phylogeny.fr interface.
The phylogenetic tree was constructed by PhyML using the approximate
likelihood-ratio test and using the
substitution model of Jones, Taylor, and Thornton with default parameters,
and gaps were removed from the alignment. The tree topology was confirmed
with COBALT via the National Center for Biotechnology Information
server. The sequences are identified
in the figure using the following abbreviations corresponding to the
following accession numbers: Afumig-Mn, Aspergillus
fumigatus MnSOD (Eukaryota-mito) GI:18158811; Ahydro-Fe, Aeromonas hydrophila FeSOD
(Gammaproteobacteria-Fe) GI:75530508; Anabae-Mn, Anabaena MnSOD (Cyanobacteria) GI:23200075 H2O2; Apernix-Mn/Fe, Aeropyrum pernix Mn/FeSOD (Crenarchaeota) GI:321159640; Athali-Fe, Arabidopsis
thaliana FeSOD (Viridiplantae) GI:332659609; Athal-Mn, Arabidopsis thaliana MnSOD
(Viridiplantae-mito) GI:15228407; Avine-Fe, Azotobacter vinelandii FeSOD (Gammaproteobacteria-Fe) GI:226720755 Anal.; Bthuri-Mn, Bacillus
thuringiensis MnSOD (Firmicutes) GI:228830333; Cauran-Mn, Chloroflexus aurantiacus MnSOD (Chloroflexii) GI:31074373 Anal.; Cburne-Fe, Coxiella burnetii FeSOD (Gammaproteobacteria-Fe) GI:145002 H2O2; Cgluta-Mn, Corynebacterium
glutamicum MnSOD (Actinobacteria) GI:81783000; Cjejun-Fe, Campylobacter jejuni FeSOD
(Epsilonproteobacteria) GI:218561849 H2O2; Creinh-Fe, Chlamydomonas
reinhard FeSOD (Viridiplantae) GI:158280091; Dmelan-Mn, Drosophila melanogaster MnSOD (Eukaryota-mito) GI:7302882; Dradio-Mn, Deinococcus radiodurans MnSOD (Bacteria-Deinococ) GI:32363428; Ecoli-Fe, E. coli FeSOD (Gammaproteobacteria-Fe) GI:84028734 Anal; Ecoli-Mn, E.
coli MnSOD (Gammaproteobacteria-Mn) GI:134659 Anal;, Ehist-Fe, Entamoeba histolytica FeSOD
(protozoan-Eukaryota) GI:464774 H2O2; Ggallu-Mn, Gallus
gallus MnSOD (Eukaryota-mito) GI:15419940; Hpylor-Fe, Helicobacter pylori FeSOD
(Epsilonproteobacteria) GI:190016324; Hsap-Mn, Homo sapiens MnSOD (Eukaryota-mito) GI:24987871; Livano-Mn, Listeria ivanovii MnSOD
(Firmicutes) GI:134666; Mbark-Fe, Methanosarcina barkeri FeSOD (Euryarchaeota) GI:499627762 Anal.; Methylo-Mn, Methylomonas MnSOD (Gammaproteobacteria-Mn) GI:95281 Anal; Mpalea-Fe, Marchantia paleacea FeSOD
(Viridiplantae) GI:75243372; Msativ-Fe, Medicago sativa FeSOD (Viridiplantae) GI:75248782; Msmeg-Mn, Mycobacterium smegmatis MnSOD (Actinobacteria) GI:21264517 Anal; Mthermo-Fe, Methanobacterium
thermoauto FeSOD (Euryarchaeota) GI:23200500; Mtuber-Fe, Mycobacterium tuberculosis FeSOD (Actinobacteria) GI:809164 H2O2; Nmenin-Fe, Neisseria
meningitidis FeSOD (Betaproteobacteria) GI:7226122; Naster-Mn, Nocardia asteroides MnSOD
(Actinobacteria) GI:1711453; Nostoc-Fe, Nostoc PCC7120 FeSOD (Cyanobacteria) GI:17132032; Paeroph-Mn/Fe, Pyrobaculum aerophilum Mn/FeSOD (Crenarchaeota) GI:14917043; Pborya-Fe,: Plectonema
boryanum FeSOD (Cyanobacteria) GI:1711435 Anal; Pfalc-Fe, Plasmodium
falciparum FeSOD (protozoan-Eukaryota) GI:74946757; Pfreud-FeMn, Propionibacterium
freudenreichii (shermanii) Fe/MnSOD
(Actinobacteria) GI:5542134 Anal.; Phalo-Fe, Pseudoalteromonas haloplanktis FeSOD (Gammaproteobacteria-Fe) GI:306440524; Pleiog-Fe, Photobacterium leiognathi FeSOD (Gammaproteobacteria-Fe) GI:134643 Anal; Poliv-Mn, Paralichthys olivaceus MnSOD (Eukaryota-mito) GI:134676; Poval-Fe, Pseudomonas ovalis FeSOD (Gammaproteobacteria-Fe) GI:12084342 Anal; Ppinas-Fe, Pinus
pinaster FeSOD (Viridiplantae) GI:75223482; Scere-Mn, Saccharomyces cerevisiae MnSOD (Eukaryota-mito) GI:217035334; Ssolfa-Fe, Sulfolobus solfataricus FeSOD (Crenarchaeota) GI:14286093 Anal.;, Synech-Fe, Synechocystis 6803 FeSOD (Cyanobacteria) GI:1653111; Taest-Mn, Triticum aestivum MnSOD (Viridiplantae-mito) GI:62131095; Taq-Mn, Thermus aquaticus MnSOD
(Bacteria-Deinococ) GI:1711455; Tbruce-Fe, Trypanosoma brucei B2 FeSOD (protozoan-Eukaryota) GI:70834946 H2O2; Telong-Fe, Thermosynechococcus elongatus FeSOD (Cyanobacteria) GI:34810955; Tgondi-Fe, Toxoplasma gondii FeSOD
(protozoan-Eukaryota) GI:122066229; Vcart-Fe, Volvox carteri FeSOD (Viridiplantae) GI:121077704; Vchol-Mn, Vibrio cholerae MnSOD
(Gammaproteobacteria-Mn) GI:14039308 upregulation in absence
of Fe; Vungui-Fe, Vigna
unguiculata FeSOD (Viridiplantae) GI:56554197 H2O2; Xcamp-Mn, Xanthomonas campestris MnSOD (Gammaproteobacteria-Mn) GI:76364224.