MALDI imaging mass spectrometry: spatial molecular analysis to enable a new age of discovery
- PMID: 24686089
- PMCID: PMC4104210
- DOI: 10.1016/j.jprot.2014.03.021
MALDI imaging mass spectrometry: spatial molecular analysis to enable a new age of discovery
Abstract
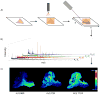
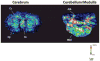
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) combines the sensitivity and selectivity of mass spectrometry with spatial analysis to provide a new dimension for histological analyses to provide unbiased visualization of the arrangement of biomolecules in tissue. As such, MALDI IMS has the capability to become a powerful new molecular technology for the biological and clinical sciences. In this review, we briefly describe several applications of MALDI IMS covering a range of molecular weights, from drugs to proteins. Current limitations and challenges are discussed along with recent developments to address these issues. This article is part of a Special Issue entitled: 20years of Proteomics in memory of Viatliano Pallini. Guest Editors: Luca Bini, Juan J. Calvete, Natacha Turck, Denis Hochstrasser and Jean-Charles Sanchez.
Keywords: MALDI imaging mass spectrometry.
Copyright © 2014 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures




References
-
- de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Froehlich F, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–U60. - PubMed
-
- Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Analytical Chemistry. 1997;69:4751–60. - PubMed
-
- Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nature Medicine. 2001;7:493–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

