Solenopsis invicta virus 3: mapping of structural proteins, ribosomal frameshifting, and similarities to Acyrthosiphon pisum virus and Kelp fly virus
- PMID: 24686475
- PMCID: PMC3970965
- DOI: 10.1371/journal.pone.0093497
Solenopsis invicta virus 3: mapping of structural proteins, ribosomal frameshifting, and similarities to Acyrthosiphon pisum virus and Kelp fly virus
Abstract
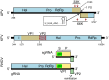
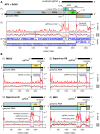
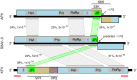
Solenopsis invicta virus 3 (SINV-3) is a positive-sense single-stranded RNA virus that infects the red imported fire ant, Solenopsis invicta. We show that the second open reading frame (ORF) of the dicistronic genome is expressed via a frameshifting mechanism and that the sequences encoding the structural proteins map to both ORF2 and the 3' end of ORF1, downstream of the sequence that encodes the RNA-dependent RNA polymerase. The genome organization and structural protein expression strategy resemble those of Acyrthosiphon pisum virus (APV), an aphid virus. The capsid protein that is encoded by the 3' end of ORF1 in SINV-3 and APV is predicted to have a jelly-roll fold similar to the capsid proteins of picornaviruses and caliciviruses. The capsid-extension protein that is produced by frameshifting, includes the jelly-roll fold domain encoded by ORF1 as its N-terminus, while the C-terminus encoded by the 5' half of ORF2 has no clear homology with other viral structural proteins. A third protein, encoded by the 3' half of ORF2, is associated with purified virions at sub-stoichiometric ratios. Although the structural proteins can be translated from the genomic RNA, we show that SINV-3 also produces a subgenomic RNA encoding the structural proteins. Circumstantial evidence suggests that APV may also produce such a subgenomic RNA. Both SINV-3 and APV are unclassified picorna-like viruses distantly related to members of the order Picornavirales and the family Caliciviridae. Within this grouping, features of the genome organization and capsid domain structure of SINV-3 and APV appear more similar to caliciviruses, perhaps suggesting the basis for a "Calicivirales" order.
Conflict of interest statement
Figures


References
-
- van den Heuvel JF, Hummelen H, Verbeek M, Dullemans AM, van der Wilk F (1997) Characteristics of Acyrthosiphon pisum virus, a newly identified virus infecting the pea aphid. J Invertebr Pathol 70: 169–176. - PubMed
-
- van der Wilk F, Dullemans AM, Verbeek M, Van den Heuvel JF (1997) Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology 238: 353–362. - PubMed
-
- Scotti PD, Gibbs AJ, Wrigley NG (1976) Kelp fly virus. J Gen Virol 30: 1–9.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

