Acidosis overrides oxygen deprivation to maintain mitochondrial function and cell survival
- PMID: 24686499
- PMCID: PMC3988820
- DOI: 10.1038/ncomms4550
Acidosis overrides oxygen deprivation to maintain mitochondrial function and cell survival
Abstract
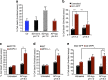
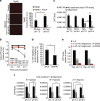
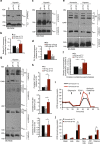
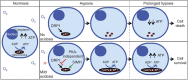
Sustained cellular function and viability of high-energy demanding post-mitotic cells rely on the continuous supply of ATP. The utilization of mitochondrial oxidative phosphorylation for efficient ATP generation is a function of oxygen levels. As such, oxygen deprivation, in physiological or pathological settings, has profound effects on cell metabolism and survival. Here we show that mild extracellular acidosis, a physiological consequence of anaerobic metabolism, can reprogramme the mitochondrial metabolic pathway to preserve efficient ATP production regardless of oxygen levels. Acidosis initiates a rapid and reversible homeostatic programme that restructures mitochondria, by regulating mitochondrial dynamics and cristae architecture, to reconfigure mitochondrial efficiency, maintain mitochondrial function and cell survival. Preventing mitochondrial remodelling results in mitochondrial dysfunction, fragmentation and cell death. Our findings challenge the notion that oxygen availability is a key limiting factor in oxidative metabolism and brings forth the concept that mitochondrial morphology can dictate the bioenergetic status of post-mitotic cells.
Figures
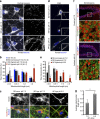

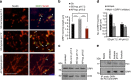
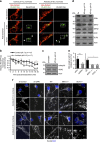
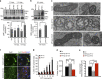
References
-
- Lin M. T. & Beal M. F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 (2006). - PubMed
-
- Newmeyer D. D. & Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112, 481–490 (2003). - PubMed
-
- Detmer S. A. & Chan D. C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8, 870–879 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

