Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling
- PMID: 24686847
- PMCID: PMC4004585
- DOI: 10.1038/ng.2933
Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling
Abstract
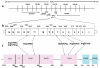
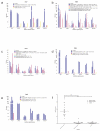
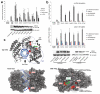
The type I interferon system is integral to human antiviral immunity. However, inappropriate stimulation or defective negative regulation of this system can lead to inflammatory disease. We sought to determine the molecular basis of genetically uncharacterized cases of the type I interferonopathy Aicardi-Goutières syndrome and of other undefined neurological and immunological phenotypes also demonstrating an upregulated type I interferon response. We found that heterozygous mutations in the cytosolic double-stranded RNA receptor gene IFIH1 (also called MDA5) cause a spectrum of neuroimmunological features consistently associated with an enhanced interferon state. Cellular and biochemical assays indicate that these mutations confer gain of function such that mutant IFIH1 binds RNA more avidly, leading to increased baseline and ligand-induced interferon signaling. Our results demonstrate that aberrant sensing of nucleic acids can cause immune upregulation.
Figures

Comment in
-
MDA5 and autoimmune disease.Nat Genet. 2014 May;46(5):418-9. doi: 10.1038/ng.2959. Nat Genet. 2014. PMID: 24769716 No abstract available.
-
Autosomal dominant IFIH1 gain-of-function mutations cause Aicardi-Goutières syndrome.Clin Genet. 2014 Nov;86(5):473-4. doi: 10.1111/cge.12471. Epub 2014 Sep 8. Clin Genet. 2014. PMID: 25080300
References
-
- Gresser I, Tovey MG, Maury C, Chouroulinkov I. Lethality of interferon preparations for newborn mice. Nature. 1975;258:76–8. - PubMed
-
- Gresser I, et al. Progressive glomerulonephritis in mice treated with interferon preparations at birth. Nature. 1976;263:420–2. - PubMed
-
- Gresser I, et al. Interferon-induced disease in mice and rats. Ann N Y Acad Sci. 1980;350:12–20. - PubMed
-
- Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–8. - PubMed
-
- Crow YJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–20. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- HL-102924/HL/NHLBI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- HL-102925/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- HL-102923/HL/NHLBI NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102926/HL/NHLBI NIH HHS/United States
- UC2 HL103010/HL/NHLBI NIH HHS/United States
- HL-103010/HL/NHLBI NIH HHS/United States
- HL-102926/HL/NHLBI NIH HHS/United States
- 309449/ERC_/European Research Council/International
- UC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102924/HL/NHLBI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UC2 HL102925/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

