Discovery of colorectal cancer biomarker candidates by membrane proteomic analysis and subsequent verification using selected reaction monitoring (SRM) and tissue microarray (TMA) analysis
- PMID: 24687888
- PMCID: PMC4047467
- DOI: 10.1074/mcp.M113.037093
Discovery of colorectal cancer biomarker candidates by membrane proteomic analysis and subsequent verification using selected reaction monitoring (SRM) and tissue microarray (TMA) analysis
Abstract
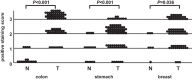
Recent advances in quantitative proteomic technology have enabled the large-scale validation of biomarkers. We here performed a quantitative proteomic analysis of membrane fractions from colorectal cancer tissue to discover biomarker candidates, and then extensively validated the candidate proteins identified. A total of 5566 proteins were identified in six tissue samples, each of which was obtained from polyps and cancer with and without metastasis. GO cellular component analysis predicted that 3087 of these proteins were membrane proteins, whereas TMHMM algorithm predicted that 1567 proteins had a transmembrane domain. Differences were observed in the expression of 159 membrane proteins and 55 extracellular proteins between polyps and cancer without metastasis, while the expression of 32 membrane proteins and 17 extracellular proteins differed between cancer with and without metastasis. A total of 105 of these biomarker candidates were quantitated using selected (or multiple) reaction monitoring (SRM/MRM) with stable synthetic isotope-labeled peptides as an internal control. The results obtained revealed differences in the expression of 69 of these proteins, and this was subsequently verified in an independent set of patient samples (polyps (n = 10), cancer without metastasis (n = 10), cancer with metastasis (n = 10)). Significant differences were observed in the expression of 44 of these proteins, including ITGA5, GPRC5A, PDGFRB, and TFRC, which have already been shown to be overexpressed in colorectal cancer, as well as proteins with unknown function, such as C8orf55. The expression of C8orf55 was also shown to be high not only in colorectal cancer, but also in several cancer tissues using a multicancer tissue microarray, which included 1150 cores from 14 cancer tissues. This is the largest verification study of biomarker candidate membrane proteins to date; our methods for biomarker discovery and subsequent validation using SRM/MRM will contribute to the identification of useful biomarker candidates for various cancers. Data are available via ProteomeXchange with identifier PXD000851.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents, Mol. Cell. Proteomics 3, 1154–1169 - PubMed
-
- Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 - PubMed
-
- Whiteaker J. R., Zhang H., Zhao L., Wang P., Kelly-Spratt K. S., Ivey R. G., Piening B. D., Feng L. C., Kasarda E., Gurley K. E., Eng J. K., Chodosh L. A., Kemp C. J., McIntosh M. W., Paulovich A. G. (2007) Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res. 6, 3962–3975 - PubMed
-
- Whiteaker J. R., Lin C., Kennedy J., Hou L., Trute M., Sokal I., Yan P., Schoenherr R. M., Zhao L., Voytovich U. J., Kelly-Spratt K. S., Krasnoselsky A., Gafken P. R., Hogan J. M., Jones L. A., Wang P., Amon L., Chodosh L. A., Nelson P. S., McIntosh M. W., Kemp C. J., Paulovich A. G. (2011) A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat. Biotechnol. 29, 625–634 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

