Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products
- PMID: 24688664
- PMCID: PMC3962153
- DOI: 10.5936/csbj.201210004
Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products
Abstract
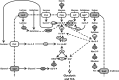
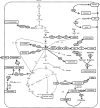
Corynebacterium glutamicum is well known as the amino acid-producing workhorse of fermentation industry, being used for multi-million-ton scale production of glutamate and lysine for more than 60 years. However, it is only recently that extensive research has focused on engineering it beyond the scope of amino acids. Meanwhile, a variety of corynebacterial strains allows access to alternative carbon sources and/or allows production of a wide range of industrially relevant compounds. Some of these efforts set new standards in terms of titers and productivities achieved whereas others represent a proof-of-principle. These achievements manifest the position of C. glutamicum as an important industrial microorganism with capabilities far beyond the traditional amino acid production. In this review we focus on the state of the art of metabolic engineering of C. glutamicum for utilization of alternative carbon sources, (e.g. coming from wastes and unprocessed sources), and construction of C. glutamicum strains for production of new products such as diamines, organic acids and alcohols.
Keywords: amino acids; diamines; glycerol; hemicellulose; organic acids.
Figures
Similar articles
-
Synthetic biology approaches to access renewable carbon source utilization in Corynebacterium glutamicum.Appl Microbiol Biotechnol. 2018 Nov;102(22):9517-9529. doi: 10.1007/s00253-018-9358-x. Epub 2018 Sep 14. Appl Microbiol Biotechnol. 2018. PMID: 30218378 Review.
-
Engineering microbial cell factories: Metabolic engineering of Corynebacterium glutamicum with a focus on non-natural products.Biotechnol J. 2015 Aug;10(8):1170-84. doi: 10.1002/biot.201400590. Epub 2015 Jul 24. Biotechnol J. 2015. PMID: 26216246 Review.
-
Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery.Appl Microbiol Biotechnol. 2018 May;102(9):3915-3937. doi: 10.1007/s00253-018-8896-6. Epub 2018 Mar 20. Appl Microbiol Biotechnol. 2018. PMID: 29557518 Review.
-
Microbial Production of Amino Acid-Related Compounds.Adv Biochem Eng Biotechnol. 2017;159:255-269. doi: 10.1007/10_2016_34. Adv Biochem Eng Biotechnol. 2017. PMID: 27872963 Review.
-
Coproduction of cell-bound and secreted value-added compounds: Simultaneous production of carotenoids and amino acids by Corynebacterium glutamicum.Bioresour Technol. 2018 Jan;247:744-752. doi: 10.1016/j.biortech.2017.09.167. Epub 2017 Sep 27. Bioresour Technol. 2018. PMID: 30060409
Cited by
-
High-efficiency production of the antimicrobial peptide pediocin PA-1 in metabolically engineered Corynebacterium glutamicum using a microaerobic process at acidic pH and elevated levels of bivalent calcium ions.Microb Cell Fact. 2023 Feb 27;22(1):41. doi: 10.1186/s12934-023-02044-y. Microb Cell Fact. 2023. PMID: 36849884 Free PMC article.
-
Metabolic engineering of Corynebacterium glutamicum for methanol metabolism.Appl Environ Microbiol. 2015 Mar;81(6):2215-25. doi: 10.1128/AEM.03110-14. Epub 2015 Jan 16. Appl Environ Microbiol. 2015. PMID: 25595770 Free PMC article.
-
Review of the Proteomics and Metabolic Properties of Corynebacterium glutamicum.Microorganisms. 2024 Aug 15;12(8):1681. doi: 10.3390/microorganisms12081681. Microorganisms. 2024. PMID: 39203523 Free PMC article. Review.
-
Crossing boundaries: the importance of cellular membranes in industrial biotechnology.J Ind Microbiol Biotechnol. 2017 May;44(4-5):721-733. doi: 10.1007/s10295-016-1858-z. Epub 2016 Nov 11. J Ind Microbiol Biotechnol. 2017. PMID: 27837352 Review.
-
Updates on industrial production of amino acids using Corynebacterium glutamicum.World J Microbiol Biotechnol. 2016 Jun;32(6):105. doi: 10.1007/s11274-016-2060-1. Epub 2016 Apr 27. World J Microbiol Biotechnol. 2016. PMID: 27116971 Review.
References
-
- Kinoshita S, Udaka S, Shimono M (1957) Studies on the amino acid fermentation. Production of L-glutamic acid by various microorganisms. J Gen Appl Microbiol 3: 193–205 - PubMed
-
- Ajinomoto (2010) Food Products Business. Available: http://www.ajinomoto.com/ir/pdf/Food-Oct2010.pdf.
-
- Ajinomoto (2011) Feed-Use Amino Acids Business. Available: http://www.ajinomoto.com/ir/pdf/Feed-useAA-Oct2011.pdf
-
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, et al. (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol 104: 5–25 - PubMed
-
- Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62: 99–109 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
