Metabolic modelling in the development of cell factories by synthetic biology
- PMID: 24688669
- PMCID: PMC3962133
- DOI: 10.5936/csbj.201210009
Metabolic modelling in the development of cell factories by synthetic biology
Abstract
Cell factories are commonly microbial organisms utilized for bioconversion of renewable resources to bulk or high value chemicals. Introduction of novel production pathways in chassis strains is the core of the development of cell factories by synthetic biology. Synthetic biology aims to create novel biological functions and systems not found in nature by combining biology with engineering. The workflow of the development of novel cell factories with synthetic biology is ideally linear which will be attainable with the quantitative engineering approach, high-quality predictive models, and libraries of well-characterized parts. Different types of metabolic models, mathematical representations of metabolism and its components, enzymes and metabolites, are useful in particular phases of the synthetic biology workflow. In this minireview, the role of metabolic modelling in synthetic biology will be discussed with a review of current status of compatible methods and models for the in silico design and quantitative evaluation of a cell factory.
Keywords: chassis; constraint-based; flux; kinetics; simulation.
Figures
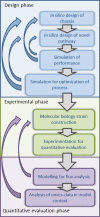
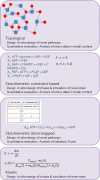
Similar articles
-
Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts.Biotechnol Adv. 2021 Mar-Apr;47:107695. doi: 10.1016/j.biotechadv.2021.107695. Epub 2021 Jan 16. Biotechnol Adv. 2021. PMID: 33465474 Review.
-
Kluyveromyces as promising yeast cell factories for industrial bioproduction: From bio-functional design to applications.Biotechnol Adv. 2023 May-Jun;64:108125. doi: 10.1016/j.biotechadv.2023.108125. Epub 2023 Mar 2. Biotechnol Adv. 2023. PMID: 36870581 Review.
-
Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production.Chem Soc Rev. 2020 Jul 21;49(14):4615-4636. doi: 10.1039/d0cs00155d. Chem Soc Rev. 2020. PMID: 32567619 Review.
-
Application of synthetic biology for production of chemicals in yeast Saccharomyces cerevisiae.FEMS Yeast Res. 2015 Feb;15(1):1-12. doi: 10.1111/1567-1364.12213. Epub 2015 Jan 14. FEMS Yeast Res. 2015. PMID: 25238571
-
Design and construction of microbial cell factories based on systems biology.Synth Syst Biotechnol. 2022 Nov 18;8(1):176-185. doi: 10.1016/j.synbio.2022.11.001. eCollection 2023 Mar. Synth Syst Biotechnol. 2022. PMID: 36874510 Free PMC article. Review.
Cited by
-
In silico gene knockout prediction using a hybrid of Bat algorithm and minimization of metabolic adjustment.J Integr Bioinform. 2021 Aug 4;18(3):20200037. doi: 10.1515/jib-2020-0037. J Integr Bioinform. 2021. PMID: 34348418 Free PMC article.
-
Shikimic Acid Production in Escherichia coli: From Classical Metabolic Engineering Strategies to Omics Applied to Improve Its Production.Front Bioeng Biotechnol. 2015 Sep 23;3:145. doi: 10.3389/fbioe.2015.00145. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 26442259 Free PMC article. Review.
-
Metabolic modeling and response surface analysis of an Escherichia coli strain engineered for shikimic acid production.BMC Syst Biol. 2018 Nov 12;12(1):102. doi: 10.1186/s12918-018-0632-4. BMC Syst Biol. 2018. PMID: 30419897 Free PMC article.
-
Escherichia coli redox mutants as microbial cell factories for the synthesis of reduced biochemicals.Comput Struct Biotechnol J. 2013 Jan 18;3:e201210019. doi: 10.5936/csbj.201210019. eCollection 2012. Comput Struct Biotechnol J. 2013. PMID: 24688679 Free PMC article. Review.
-
Machine learning-assisted medium optimization revealed the discriminated strategies for improved production of the foreign and native metabolites.Comput Struct Biotechnol J. 2023 Apr 20;21:2654-2663. doi: 10.1016/j.csbj.2023.04.020. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37138901 Free PMC article.
References
-
- Nielsen J, Keasling JD (2011) Synergies between synthetic biology and metabolic engineering. Nat Biotechnol 29: 693–695 - PubMed
-
- Varma A, Palsson B (1994) Metabolic flux balancing: basic concepts, scientific and practical use. Nat Biotechnol 12: 994–998
-
- Price N, Papin J, Schilling C, Palsson B (2003) Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol 21: 162–169 - PubMed
-
- Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, Nielsen J (2009) Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng 11: 328–334 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
