Escherichia coli redox mutants as microbial cell factories for the synthesis of reduced biochemicals
- PMID: 24688679
- PMCID: PMC3962086
- DOI: 10.5936/csbj.201210019
Escherichia coli redox mutants as microbial cell factories for the synthesis of reduced biochemicals
Abstract
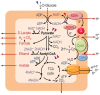
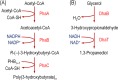
Bioprocesses conducted under conditions with restricted O2 supply are increasingly exploited for the synthesis of reduced biochemicals using different biocatalysts. The model facultative aerobe Escherichia coli, the microbial cell factory par excellence, has elaborate sensing and signal transduction mechanisms that respond to the availability of electron acceptors and alternative carbon sources in the surrounding environment. In particular, the ArcBA and CreBC two-component signal transduction systems are largely responsible for the metabolic regulation of redox control in response to O2 availability and carbon source utilization, respectively. Significant advances in the understanding of the biochemical, genetic, and physiological duties of these regulatory systems have been achieved in recent years. This situation allowed to rationally-design novel engineering approaches that ensure optimal carbon and energy flows within central metabolism, as well as to manipulate redox homeostasis, in order to optimize the production of industrially-relevant metabolites. In particular, metabolic flux analysis provided new clues to understand the metabolic regulation mediated by the ArcBA and CreBC systems. Genetic manipulation of these regulators proved useful for designing microbial cells factories tailored for the synthesis of reduced biochemicals with added value, such as poly(3-hydroxybutyrate), under conditions with restricted O2 supply. This network-wide strategy is in contrast with traditional metabolic engineering approaches, that entail direct modification of the pathway(s) at stake, and opens new avenues for the targeted modulation of central catabolic pathways at the transcriptional level.
Keywords: ArcBA; CreBC; Escherichia coli; metabolic flux analysis; polyhydroxyalkanoates; redox homeostasis; reduced biochemicals.
Figures
References
-
- Verlinden RAJ, Hill DJ, Kenward MA, Williams CD, Radecka I (2007) Bacterial synthesis of biodegradable polyhydroxyalkanoates. J Appl Microbiol 102: 1437–1449 - PubMed
-
- Gomez JGC, Méndez BS, Nikel PI, Pettinari MJ, Prieto MA, et al. (2012) Making green polymers even greener: towards sustainable production of polyhydroxyalkanoates from agroindustrial by-products In: Petre M, editor. Advances in applied biotechnology. Rijeka, Croatia: InTech; pp. 41–62
-
- Clomburg JM, Gonzalez R (2010) Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl Microbiol Biotechnol 86: 419–434 - PubMed
-
- Geddes CC, Nieves IU, Ingram LO (2011) Advances in ethanol production. Curr Opin Biotechnol 22: 312–319 - PubMed
-
- Biebl H (1999) Microbial production of 1,3-propanediol. Appl Microbiol Biotechnol 52: 289–297 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
