Of mice and men: Dissecting the interaction between Listeria monocytogenes Internalin A and E-cadherin
- PMID: 24688730
- PMCID: PMC3962206
- DOI: 10.5936/csbj.201303022
Of mice and men: Dissecting the interaction between Listeria monocytogenes Internalin A and E-cadherin
Abstract
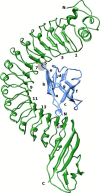
We report a study of the interaction between internalin A (inlA) and human or murine E-cadherin (Ecad). inlA is used by Listeria monocytogenes to internalize itself into host cell, but the bacterium is unable to invade murine cells, which has been attributed to the difference in sequence between hEcad and mEcad. Using molecular dynamics simulations, MM/GBSA free energy calculations, hydrogen bond analysis, water characterization and umbrella sampling, we provide a complete atomistic picture of the binding between inlA and Ecad. We dissect key residues in the protein-protein interface and analyze the energetics using MM/GBSA. From this analysis it is clear that the binding of inlA-mEcad is weaker than inlA-hEcad, on par with the experimentally observed inability of inlA to bind to mEcad. However, extended MD simulations of 200 ns in length show no destabilization of the inlA-mEcad complex and the estimation of the potential of mean force (PMF) using umbrella sampling corroborates this conclusion. The binding strength computed from the PMFs show no significant difference between the two protein complexes. Hence, our study suggests that the inability of L. monocytogenes to invade murine cells cannot be explained by processes at the nanosecond to sub-microsecond time scale probed by the simulations performed here.
Keywords: E-cadherin; Internalin A; energy decomposition; molecular dynamics; umbrella sampling.
Figures







References
-
- Hamon M, Bierne H, Cossart P (2006) Listeria monocytogenes: a multifaceted model. Nat rev Microbiol 4: 423–434 - PubMed
-
- Bierne H, Sabet C, Personnic N, Cossart P (2007) Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect 9: 1156–1166 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
