Regulation of gene expression in the genomic context
- PMID: 24688749
- PMCID: PMC3962188
- DOI: 10.5936/csbj.201401001
Regulation of gene expression in the genomic context
Abstract
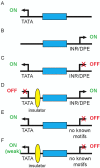
Metazoan life is dependent on the proper temporal and spatial control of gene expression within the many cells-essentially all with the identical genome-that make up the organism. While much is understood about how individual gene regulatory elements function, many questions remain about how they interact to maintain correct regulation globally throughout the genome. In this review we summarize the basic features and functions of the crucial regulatory elements promoters, enhancers, and insulators and discuss some of the ways in which proper interactions between these elements is realized. We focus in particular on the role of core promoter sequences and propose explanations for some of the contradictory results seen in experiments aimed at understanding insulator function. We suggest that gene regulation depends on local genomic context and argue that more holistic in vivo investigations that take into account multiple local features will be necessary to understand how genome-wide gene regulation is maintained.
Figures
Similar articles
-
A comprehensive map of insulator elements for the Drosophila genome.PLoS Genet. 2010 Jan 15;6(1):e1000814. doi: 10.1371/journal.pgen.1000814. PLoS Genet. 2010. PMID: 20084099 Free PMC article.
-
Genome-Wide Mapping Targets of the Metazoan Chromatin Remodeling Factor NURF Reveals Nucleosome Remodeling at Enhancers, Core Promoters and Gene Insulators.PLoS Genet. 2016 Apr 5;12(4):e1005969. doi: 10.1371/journal.pgen.1005969. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27046080 Free PMC article.
-
Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection.Genome Biol. 2020 Mar 23;21(1):75. doi: 10.1186/s13059-020-01984-7. Genome Biol. 2020. PMID: 32293525 Free PMC article.
-
Mechanisms of insulator function in gene regulation and genomic imprinting.Int Rev Cytol. 2003;232:89-127. doi: 10.1016/s0074-7696(03)32003-0. Int Rev Cytol. 2003. PMID: 14711117 Review.
-
Altered TERT promoter and other genomic regulatory elements: occurrence and impact.Int J Cancer. 2017 Sep 1;141(5):867-876. doi: 10.1002/ijc.30735. Epub 2017 Apr 29. Int J Cancer. 2017. PMID: 28407294 Review.
Cited by
-
Histone H4 lysine 20 monomethylation is not a mark of transcriptional silencers.bioRxiv [Preprint]. 2025 Jan 13:2025.01.09.632211. doi: 10.1101/2025.01.09.632211. bioRxiv. 2025. PMID: 39868205 Free PMC article. Preprint.
-
Homotypic clusters of transcription factor binding sites: A model system for understanding the physical mechanics of gene expression.Comput Struct Biotechnol J. 2014 Aug 1;10(17):63-9. doi: 10.1016/j.csbj.2014.07.005. eCollection 2014 Jul. Comput Struct Biotechnol J. 2014. PMID: 25349675 Free PMC article. Review.
-
Formation and Development of Taproots in Deciduous Tree Species.Front Plant Sci. 2021 Dec 2;12:772567. doi: 10.3389/fpls.2021.772567. eCollection 2021. Front Plant Sci. 2021. PMID: 34925417 Free PMC article. Review.
-
Analysis of Association of Angiotensin II Type 1 Receptor Gene A1166C Gene Polymorphism with Essential Hypertension.Indian J Clin Biochem. 2018 Jan;33(1):53-60. doi: 10.1007/s12291-017-0644-7. Epub 2017 Mar 13. Indian J Clin Biochem. 2018. PMID: 29371770 Free PMC article.
-
An explainable artificial intelligence approach for decoding the enhancer histone modifications code and identification of novel enhancers in Drosophila.Genome Biol. 2021 Nov 8;22(1):308. doi: 10.1186/s13059-021-02532-7. Genome Biol. 2021. PMID: 34749786 Free PMC article.
References
-
- Nestorov P, Tardat M, Peters AH (2013) H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr Top Dev Biol 104: 243–291 - PubMed
-
- Smale ST, Kadonaga JT (2003) THE RNA POLYMERASE II CORE PROMOTER. Annual Review of Biochemistry 72: 449–479 - PubMed
-
- Juven-Gershon T, Hsu JY, Kadonaga JT (2006) Perspectives on the RNA polymerase II core promoter. Biochem Soc Trans 34: 1047–1050 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
