Integrative analysis of miRNA-mRNA and miRNA-miRNA interactions
- PMID: 24689063
- PMCID: PMC3945032
- DOI: 10.1155/2014/907420
Integrative analysis of miRNA-mRNA and miRNA-miRNA interactions
Abstract
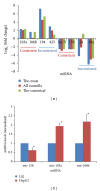
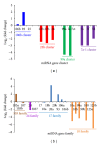
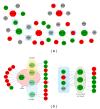
MicroRNAs (miRNAs) are small, noncoding regulatory molecules. They are involved in many essential biological processes and act by suppressing gene expression. The present work reports an integrative analysis of miRNA-mRNA and miRNA-miRNA interactions and their regulatory patterns using high-throughput miRNA and mRNA datasets. Aberrantly expressed miRNA and mRNA profiles were obtained based on fold change analysis, and qRT-PCR was used for further validation of deregulated miRNAs. miRNAs and target mRNAs were found to show various expression patterns. miRNA-miRNA interactions and clustered/homologous miRNAs were also found to contribute to the flexible and selective regulatory network. Interacting miRNAs (e.g., miRNA-103a and miR-103b) showed more pronounced differences in expression, which suggests the potential "restricted interaction" in the miRNA world. miRNAs from the same gene clusters (e.g., miR-23b gene cluster) or gene families (e.g., miR-10 gene family) always showed the same types of deregulation patterns, although they sometimes differed in expression levels. These clustered and homologous miRNAs may have close functional relationships, which may indicate collaborative interactions between miRNAs. The integrative analysis of miRNA-mRNA based on biological characteristics of miRNA will further enrich miRNA study.
Figures
References
-
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature Reviews Genetics. 2011;12(2):99–110. - PubMed
-
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. The Journal of Biological Chemistry. 2008;283(2):1026–1033. - PubMed
-
- Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124(6):1169–1181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

