Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content in Arabidopsis leaves in response to microbial products
- PMID: 24690446
- PMCID: PMC4021374
- DOI: 10.1186/1471-2229-14-84
Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content in Arabidopsis leaves in response to microbial products
Abstract
Background: The production and use of biologically derived soil additives is one of the fastest growing sectors of the fertilizer industry. These products have been shown to improve crop yields while at the same time reducing fertilizer inputs to and nutrient loss from cropland. The mechanisms driving the changes in primary productivity and soil processes are poorly understood and little is known about changes in secondary productivity associated with the use of microbial products. Here we investigate secondary metabolic responses to a biologically derived soil additive by monitoring changes in the phenlypropanoid (PP) pathway in Arabidopsis thaliana.
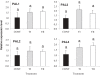
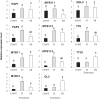
Results: This study was designed to test the influence of one of these products (Soil Builder™-AF, SB) on secondary metabolism after being applied at different times. One time (TI) application of SB to Arabidopsis increased the accumulation of flavonoids compared to multiple (TII) applications of the same products. Fourteen phenolic compounds including flavonols and anothocyanins were identified by mass spectrometry. Kaempferol-3,7-O-bis-α-L-rhamnoside and quercetin 3,7-dirhamnoside, the major compounds, increased 3-fold and 4-fold, respectively compared to control in the TI treatment. The most abundant anthocyanin was cyanidin 3-rhamnoglucoside, which increased 3-fold and 2-fold in TI compared to the control and TII, respectively. Simultaneously, the expression of genes coding for key enzymes in the PP pathway (phenylalanine ammonia lyase, cinnamate 4-hydroxylase, chalcone synthase, flavonoid-3'-O-hydroxylase, flavonol synthase1 and dihydroflavonol-4-reductase) and regulatory genes (production of anthocyanin pigment2, MYB12, MYB113, MYB114, EGL3, and TT8) were up-regulated in both treatments (TI and TII). Furthermore, application of TI and TII induced expression of the lignin pathway genes (hydroxyl cinamyl transferase, caffeyl-CoA O-methyl transferase, cinnamyl alcohol dehydrogenase, cinnamyl-CoA reductase, secondary wall-associated NAC domain protein1, MYB58 and MYB63 resulting in higher accumulation of lignin content compared to the control.
Conclusions: These results indicate that the additions of microbially based soil additives have a perceptible influence on phenylpropanoid pathway gene regulation and its production of secondary metabolites. These findings open an avenue of research to investigate the mode of action of microbially-based soil additives which may assist in the sustainable production of food, feed, fuel and fiber.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

