DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns
- PMID: 24690455
- PMCID: PMC4053947
- DOI: 10.1186/gb-2014-15-4-r54
DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns
Erratum in
-
Erratum to: DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns.Genome Biol. 2016 Nov 1;17(1):224. doi: 10.1186/s13059-016-1091-0. Genome Biol. 2016. PMID: 27802833 Free PMC article. No abstract available.
Abstract
Background: DNA epigenetic modifications, such as methylation, are important regulators of tissue differentiation, contributing to processes of both development and cancer. Profiling the tissue-specific DNA methylome patterns will provide novel insights into normal and pathogenic mechanisms, as well as help in future epigenetic therapies. In this study, 17 somatic tissues from four autopsied humans were subjected to functional genome analysis using the Illumina Infinium HumanMethylation450 BeadChip, covering 486 428 CpG sites.
Results: Only 2% of the CpGs analyzed are hypermethylated in all 17 tissue specimens; these permanently methylated CpG sites are located predominantly in gene-body regions. In contrast, 15% of the CpGs are hypomethylated in all specimens and are primarily located in regions proximal to transcription start sites. A vast number of tissue-specific differentially methylated regions are identified and considered likely mediators of tissue-specific gene regulatory mechanisms since the hypomethylated regions are closely related to known functions of the corresponding tissue. Finally, a clear inverse correlation is observed between promoter methylation within CpG islands and gene expression data obtained from publicly available databases.
Conclusions: This genome-wide methylation profiling study identified tissue-specific differentially methylated regions in 17 human somatic tissues. Many of the genes corresponding to these differentially methylated regions contribute to tissue-specific functions. Future studies may use these data as a reference to identify markers of perturbed differentiation and disease-related pathogenic mechanisms.
Figures
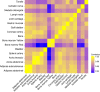
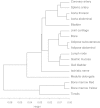

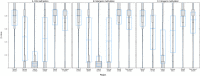

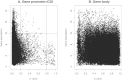
 ), bone (
), bone ( ), bone marrows (
), bone marrows ( ), lymph node (
), lymph node ( ), medulla oblongata (+), and tonsils (×).
), medulla oblongata (+), and tonsils (×).References
-
- Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–340. doi: 10.1097/EDE.0b013e31824523f0. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

