Myogenic program dysregulation is contributory to disease pathogenesis in spinal muscular atrophy
- PMID: 24691550
- PMCID: PMC4103674
- DOI: 10.1093/hmg/ddu142
Myogenic program dysregulation is contributory to disease pathogenesis in spinal muscular atrophy
Abstract
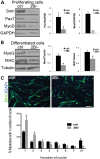
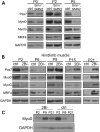
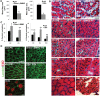
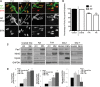
Mutations in the survival motor neuron (SMN1) gene lead to the neuromuscular disease spinal muscular atrophy (SMA). Although SMA is primarily considered as a motor neuron disease, the importance of muscle defects in its pathogenesis has not been fully examined. We use both primary cell culture and two different SMA model mice to demonstrate that reduced levels of Smn lead to a profound disruption in the expression of myogenic genes. This disruption was associated with a decrease in myofiber size and an increase in immature myofibers, suggesting that Smn is crucial for myogenic gene regulation and early muscle development. Histone deacetylase inhibitor trichostatin A treatment of SMA model mice increased myofiber size, myofiber maturity and attenuated the disruption of the myogenic program in these mice. Taken together, our work highlights the important contribution of myogenic program dysregulation to the muscle weakness observed in SMA.
© The Author 2014. Published by Oxford University Press.
Figures
References
-
- Boyer J.G., Bowerman M., Kothary R. The many faces of SMN: deciphering the function critical to spinal muscular atrophy pathogenesis. Future Neurol. 2010;5:873–890.
-
- Hahnen E., Forkert R., Marke C., Rudnik-Schoneborn S., Schonling J., Zerres K., Wirth B. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum. Mol. Genet. 1995;4:1927–1933. - PubMed
-
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Rochette C.F., Gilbert N., Simard L.R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001;108:255–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

