The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes
- PMID: 24691961
- PMCID: PMC3970416
- DOI: 10.1101/cshperspect.a016188
The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes
Abstract
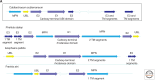
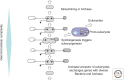
The ancestral set of eukaryotic genes is a chimera composed of genes of archaeal and bacterial origins thanks to the endosymbiosis event that gave rise to the mitochondria and apparently antedated the last common ancestor of the extant eukaryotes. The proto-mitochondrial endosymbiont is confidently identified as an α-proteobacterium. In contrast, the archaeal ancestor of eukaryotes remains elusive, although evidence is accumulating that it could have belonged to a deep lineage within the TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota) superphylum of the Archaea. Recent surveys of archaeal genomes show that the apparent ancestors of several key functional systems of eukaryotes, the components of the archaeal "eukaryome," such as ubiquitin signaling, RNA interference, and actin-based and tubulin-based cytoskeleton structures, are identifiable in different archaeal groups. We suggest that the archaeal ancestor of eukaryotes was a complex form, rooted deeply within the TACK superphylum, that already possessed some quintessential eukaryotic features, in particular, a cytoskeleton, and perhaps was capable of a primitive form of phagocytosis that would facilitate the engulfment of potential symbionts. This putative group of Archaea could have existed for a relatively short time before going extinct or undergoing genome streamlining, resulting in the dispersion of the eukaryome. This scenario might explain the difficulty with the identification of the archaeal ancestor of eukaryotes despite the straightforward detection of apparent ancestors to many signature eukaryotic functional systems.
Figures




References
-
- Adams DW, Errington J 2009. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653 - PubMed
-
- Aylett CH, Lowe J, Amos LA 2011. New insights into the mechanisms of cytomotive actin and tubulin filaments. Int Rev Cell Mol Biol 292: 1–71 - PubMed
-
- Blombach F, Makarova KS, Marrero J, Siebers B, Koonin EV, van der Oost J 2009. Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol Direct 4: 39. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
