Phosphoinositide-mediated oligomerization of a defensin induces cell lysis
- PMID: 24692446
- PMCID: PMC3968744
- DOI: 10.7554/eLife.01808
Phosphoinositide-mediated oligomerization of a defensin induces cell lysis
Abstract
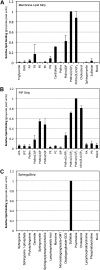
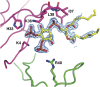
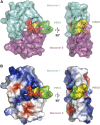
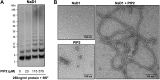
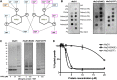
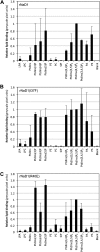
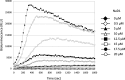
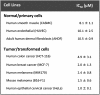
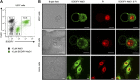
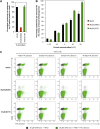
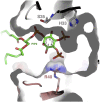
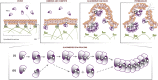
Cationic antimicrobial peptides (CAPs) such as defensins are ubiquitously found innate immune molecules that often exhibit broad activity against microbial pathogens and mammalian tumor cells. Many CAPs act at the plasma membrane of cells leading to membrane destabilization and permeabilization. In this study, we describe a novel cell lysis mechanism for fungal and tumor cells by the plant defensin NaD1 that acts via direct binding to the plasma membrane phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2). We determined the crystal structure of a NaD1:PIP2 complex, revealing a striking oligomeric arrangement comprising seven dimers of NaD1 that cooperatively bind the anionic headgroups of 14 PIP2 molecules through a unique 'cationic grip' configuration. Site-directed mutagenesis of NaD1 confirms that PIP2-mediated oligomerization is important for fungal and tumor cell permeabilization. These observations identify an innate recognition system by NaD1 for direct binding of PIP2 that permeabilizes cells via a novel membrane disrupting mechanism. DOI: http://dx.doi.org/10.7554/eLife.01808.001.
Keywords: X-ray crystallography; antimicrobial peptide; fungus; phospholipids; protein structure.
Conflict of interest statement
NLW: Head of Technology Commercialisation Hexima Ltd.
MAA: Chief Science Officer Hexima Ltd.
MDH: Vice President of Research Hexima Ltd.
The other authors declare that no competing interests exist.
Figures










References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D, Biological Crystallography 66:213–221. 10.1107/S0907444909052925 - DOI - PMC - PubMed
-
- Adda CG, Murphy VJ, Sunde M, Waddington LJ, Schloegel J, Talbo GH, Vingas K, Kienzle V, Masciantonio R, Howlett GJ, Hodder AN, Foley M, Anders RF. 2009. Plasmodium falciparum merozoite surface protein 2 is unstructured and forms amyloid-like fibrils. Molecular and Biochemical Parasitology 166:159–71. 10.1016/j.molbiopara.2009.03.012 - DOI - PMC - PubMed
-
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. 2001. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Molecular Cell 8:829–839. 10.1016/S1097-2765(01)00372-0 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

