Transcriptional analysis of apoptotic cerebellar granule neurons following rescue by gastric inhibitory polypeptide
- PMID: 24694544
- PMCID: PMC4013584
- DOI: 10.3390/ijms15045596
Transcriptional analysis of apoptotic cerebellar granule neurons following rescue by gastric inhibitory polypeptide
Abstract
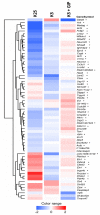
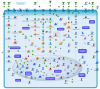
Apoptosis triggered by exogenous or endogenous stimuli is a crucial phenomenon to determine the fate of neurons, both in physiological and in pathological conditions. Our previous study established that gastric inhibitory polypeptide (Gip) is a neurotrophic factor capable of preventing apoptosis of cerebellar granule neurons (CGNs), during its pre-commitment phase. In the present study, we conducted whole-genome expression profiling to obtain a comprehensive view of the transcriptional program underlying the rescue effect of Gip in CGNs. By using DNA microarray technology, we identified 65 genes, we named survival related genes, whose expression is significantly de-regulated following Gip treatment. The expression levels of six transcripts were confirmed by real-time quantitative polymerase chain reaction. The proteins encoded by the survival related genes are functionally grouped in the following categories: signal transduction, transcription, cell cycle, chromatin remodeling, cell death, antioxidant activity, ubiquitination, metabolism and cytoskeletal organization. Our data outline that Gip supports CGNs rescue via a molecular framework, orchestrated by a wide spectrum of gene actors, which propagate survival signals and support neuronal viability.
Figures

Similar articles
-
Gastric inhibitory polypeptide and its receptor are expressed in the central nervous system and support neuronal survival.Cent Nerv Syst Agents Med Chem. 2011 Sep 1;11(3):210-22. doi: 10.2174/187152411798047771. Cent Nerv Syst Agents Med Chem. 2011. PMID: 21919873
-
Developmental expression of GPR3 in rodent cerebellar granule neurons is associated with cell survival and protects neurons from various apoptotic stimuli.Neurobiol Dis. 2014 Aug;68:215-27. doi: 10.1016/j.nbd.2014.04.007. Epub 2014 Apr 24. Neurobiol Dis. 2014. PMID: 24769160
-
Gene expression profiles of apoptotic neurons.Genomics. 2004 Sep;84(3):485-96. doi: 10.1016/j.ygeno.2004.04.006. Genomics. 2004. PMID: 15498456
-
Cerebellar granule cells as a model to study mechanisms of neuronal apoptosis or survival in vivo and in vitro.Cerebellum. 2002 Jan-Mar;1(1):41-55. doi: 10.1080/147342202753203087. Cerebellum. 2002. PMID: 12879973 Review.
-
Delineating the factors and cellular mechanisms involved in the survival of cerebellar granule neurons.Cerebellum. 2015 Jun;14(3):354-9. doi: 10.1007/s12311-015-0646-z. Cerebellum. 2015. PMID: 25596943 Review.
Cited by
-
Transcriptional landscapes at the intersection of neuronal apoptosis and substance P-induced survival: exploring pathways and drug targets.Cell Death Discov. 2016 Aug 1;2:16050. doi: 10.1038/cddiscovery.2016.50. eCollection 2016. Cell Death Discov. 2016. PMID: 27551538 Free PMC article.
-
Igf1 and Pacap rescue cerebellar granule neurons from apoptosis via a common transcriptional program.Cell Death Discov. 2015;1:15029-. doi: 10.1038/cddiscovery.2015.29. Epub 2015 Sep 7. Cell Death Discov. 2015. PMID: 26941962 Free PMC article.
-
Cracking the Code of Neuronal Cell Fate.Cells. 2023 Mar 30;12(7):1057. doi: 10.3390/cells12071057. Cells. 2023. PMID: 37048129 Free PMC article. Review.
-
Elucidating the Relationship Between Diabetes Mellitus and Parkinson's Disease Using 18F-FP-(+)-DTBZ, a Positron-Emission Tomography Probe for Vesicular Monoamine Transporter 2.Front Neurosci. 2020 Jul 14;14:682. doi: 10.3389/fnins.2020.00682. eCollection 2020. Front Neurosci. 2020. PMID: 32760240 Free PMC article. Review.
-
Altered neuronal physiology, development, and function associated with a common chromosome 15 duplication involving CHRNA7.BMC Biol. 2021 Jul 28;19(1):147. doi: 10.1186/s12915-021-01080-7. BMC Biol. 2021. PMID: 34320968 Free PMC article.
References
-
- Jellinger K.A. Challenges in neuronal apoptosis. Curr. Alzheimer Res. 2006;3:377–391. - PubMed
-
- Arends M.J., Wyllie A.H. Apoptosis: Mechanisms and roles in pathology. Internatl. Rev. Exp. Pathol. 1991;32:223–254. - PubMed
-
- Oppenheim R.W. Cell death during development of the nervous system. Annu. Rev. Neurosci. 1991;14:453–501. - PubMed
-
- Pettmann B., Henderson C.E. Neuronal cell death. Neuron. 1998;20:633–647. - PubMed
-
- Mattson M.P. Neuronal life-and-death signaling, apoptosis, and neurodegenerative disorders. Antioxid. Redox Signal. 2006;8:1997–2006. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

