Full genome virus detection in fecal samples using sensitive nucleic acid preparation, deep sequencing, and a novel iterative sequence classification algorithm
- PMID: 24695106
- PMCID: PMC3973683
- DOI: 10.1371/journal.pone.0093269
Full genome virus detection in fecal samples using sensitive nucleic acid preparation, deep sequencing, and a novel iterative sequence classification algorithm
Abstract
We have developed a full genome virus detection process that combines sensitive nucleic acid preparation optimised for virus identification in fecal material with Illumina MiSeq sequencing and a novel post-sequencing virus identification algorithm. Enriched viral nucleic acid was converted to double-stranded DNA and subjected to Illumina MiSeq sequencing. The resulting short reads were processed with a novel iterative Python algorithm SLIM for the identification of sequences with homology to known viruses. De novo assembly was then used to generate full viral genomes. The sensitivity of this process was demonstrated with a set of fecal samples from HIV-1 infected patients. A quantitative assessment of the mammalian, plant, and bacterial virus content of this compartment was generated and the deep sequencing data were sufficient to assembly 12 complete viral genomes from 6 virus families. The method detected high levels of enteropathic viruses that are normally controlled in healthy adults, but may be involved in the pathogenesis of HIV-1 infection and will provide a powerful tool for virus detection and for analyzing changes in the fecal virome associated with HIV-1 progression and pathogenesis.
Conflict of interest statement
Figures



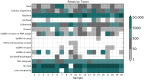
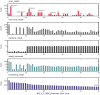

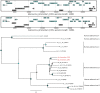

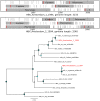



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

