Caenorhabditis elegans pathways that surveil and defend mitochondria
- PMID: 24695221
- PMCID: PMC4102179
- DOI: 10.1038/nature13204
Caenorhabditis elegans pathways that surveil and defend mitochondria
Abstract
Mitochondrial function is challenged by toxic by-products of metabolism as well as by pathogen attack. Caenorhabditis elegans normally responds to mitochondrial dysfunction with activation of mitochondrial-repair, drug-detoxification and pathogen-response pathways. Here, from a genome-wide RNA interference (RNAi) screen, we identified 45 C. elegans genes that are required to upregulate detoxification, pathogen-response and mitochondrial-repair pathways after inhibition of mitochondrial function by drug-induced or genetic disruption. Animals defective in ceramide biosynthesis are deficient in mitochondrial surveillance, and addition of particular ceramides can rescue the surveillance defects. Ceramide can also rescue the mitochondrial surveillance defects of other gene inactivations, mapping these gene activities upstream of ceramide. Inhibition of the mevalonate pathway, either by RNAi or statin drugs, also disrupts mitochondrial surveillance. Growth of C. elegans with a significant fraction of bacterial species from their natural habitat causes mitochondrial dysfunction. Other bacterial species inhibit C. elegans defence responses to a mitochondrial toxin, revealing bacterial countermeasures to animal defence.
Figures



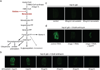
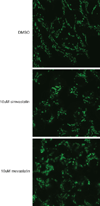
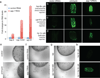

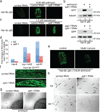
Comment in
-
Cell biology: The stressful influence of microbes.Nature. 2014 Apr 17;508(7496):328-9. doi: 10.1038/nature13220. Epub 2014 Apr 2. Nature. 2014. PMID: 24695220 No abstract available.
References
-
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Developmental cell. 2007;13:467–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

