Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers
- PMID: 24695311
- PMCID: PMC4050741
- DOI: 10.1038/nature13187
Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers
Abstract
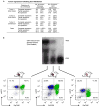
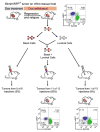
Cancer genome sequencing studies indicate that a single breast cancer typically harbours multiple genetically distinct subclones. As carcinogenesis involves a breakdown in the cell-cell cooperation that normally maintains epithelial tissue architecture, individual subclones within a malignant microenvironment are commonly depicted as self-interested competitors. Alternatively, breast cancer subclones might interact cooperatively to gain a selective growth advantage in some cases. Although interclonal cooperation has been shown to drive tumorigenesis in fruitfly models, definitive evidence for functional cooperation between epithelial tumour cell subclones in mammals is lacking. Here we use mouse models of breast cancer to show that interclonal cooperation can be essential for tumour maintenance. Aberrant expression of the secreted signalling molecule Wnt1 generates mixed-lineage mammary tumours composed of basal and luminal tumour cell subtypes, which purportedly derive from a bipotent malignant progenitor cell residing atop a tumour cell hierarchy. Using somatic Hras mutations as clonal markers, we show that some Wnt tumours indeed conform to a hierarchical configuration, but that others unexpectedly harbour genetically distinct basal Hras mutant and luminal Hras wild-type subclones. Both subclones are required for efficient tumour propagation, which strictly depends on luminally produced Wnt1. When biclonal tumours were challenged with Wnt withdrawal to simulate targeted therapy, analysis of tumour regression and relapse revealed that basal subclones recruit heterologous Wnt-producing cells to restore tumour growth. Alternatively, in the absence of a substitute Wnt source, the original subclones often evolve to rescue Wnt pathway activation and drive relapse, either by restoring cooperation or by switching to a defector strategy. Uncovering similar modes of interclonal cooperation in human cancers may inform efforts aimed at eradicating tumour cell communities.
Conflict of interest statement
The authors declare no competing financial interests.
Figures










Comment in
-
Cancer: Clonal cooperation.Nature. 2014 Apr 3;508(7494):52-3. doi: 10.1038/508052a. Nature. 2014. PMID: 24695309 No abstract available.
-
Tumour heterogeneity: A cooperative tumour cell community.Nat Rev Cancer. 2014 May;14(5):294. doi: 10.1038/nrc3732. Epub 2014 Apr 17. Nat Rev Cancer. 2014. PMID: 24739580 No abstract available.
References
-
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

