Trimmomatic: a flexible trimmer for Illumina sequence data
- PMID: 24695404
- PMCID: PMC4103590
- DOI: 10.1093/bioinformatics/btu170
Trimmomatic: a flexible trimmer for Illumina sequence data
Abstract
Motivation: Although many next-generation sequencing (NGS) read preprocessing tools already existed, we could not find any tool or combination of tools that met our requirements in terms of flexibility, correct handling of paired-end data and high performance. We have developed Trimmomatic as a more flexible and efficient preprocessing tool, which could correctly handle paired-end data.
Results: The value of NGS read preprocessing is demonstrated for both reference-based and reference-free tasks. Trimmomatic is shown to produce output that is at least competitive with, and in many cases superior to, that produced by other tools, in all scenarios tested.
Availability and implementation: Trimmomatic is licensed under GPL V3. It is cross-platform (Java 1.5+ required) and available at http://www.usadellab.org/cms/index.php?page=trimmomatic
Contact: usadel@bio1.rwth-aachen.de
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press.
Figures
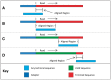

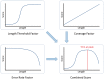
References
-
- Aronesty E. Comparison of sequencing utility programs. Open Bioinform. J. 2013;7:1–8.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

