Exploiting SNP correlations within random forest for genome-wide association studies
- PMID: 24695491
- PMCID: PMC3973686
- DOI: 10.1371/journal.pone.0093379
Exploiting SNP correlations within random forest for genome-wide association studies
Abstract
The primary goal of genome-wide association studies (GWAS) is to discover variants that could lead, in isolation or in combination, to a particular trait or disease. Standard approaches to GWAS, however, are usually based on univariate hypothesis tests and therefore can account neither for correlations due to linkage disequilibrium nor for combinations of several markers. To discover and leverage such potential multivariate interactions, we propose in this work an extension of the Random Forest algorithm tailored for structured GWAS data. In terms of risk prediction, we show empirically on several GWAS datasets that the proposed T-Trees method significantly outperforms both the original Random Forest algorithm and standard linear models, thereby suggesting the actual existence of multivariate non-linear effects due to the combinations of several SNPs. We also demonstrate that variable importances as derived from our method can help identify relevant loci. Finally, we highlight the strong impact that quality control procedures may have, both in terms of predictive power and loci identification. Variable importance results and T-Trees source code are all available at www.montefiore.ulg.ac.be/~botta/ttrees/ and github.com/0asa/TTree-source respectively.
Conflict of interest statement
Figures
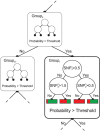
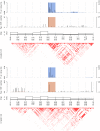
 . First row: SNP and block importances. Second row: univariate (Fisher) p-values and haplotype p-values as derived from the case/control omnibus test with
. First row: SNP and block importances. Second row: univariate (Fisher) p-values and haplotype p-values as derived from the case/control omnibus test with  degrees of freedom where
degrees of freedom where  corresponds to the number of common haplotypes (a haplotype is said to be common if its frequency is greater than
corresponds to the number of common haplotypes (a haplotype is said to be common if its frequency is greater than  in the population under study). Third row: number of haplotypes in each block. Bottom plot: ld pattern (
in the population under study). Third row: number of haplotypes in each block. Bottom plot: ld pattern ( ) in the regions.
) in the regions.References
-
- Balding DJ (2006) A tutorial on statistical methods for population association studies. Nat Rev Genet 7: 781–91. - PubMed
-
- Mccarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369. - PubMed
-
- Wang H, Misztal I, Aguilar I, Legarra A, Muir WM (2012) Genome-wide association mapping including phenotypes from relatives without genotypes. Genetics Research 94: 73–83. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

