Postnatal odorant exposure induces peripheral olfactory plasticity at the cellular level
- PMID: 24695705
- PMCID: PMC6802718
- DOI: 10.1523/JNEUROSCI.0688-13.2014
Postnatal odorant exposure induces peripheral olfactory plasticity at the cellular level
Abstract
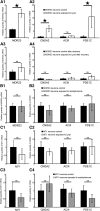
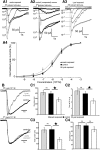
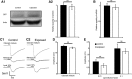
Mammalian olfactory sensory neurons (OSNs) form the primary elements of the olfactory system. Inserted in the olfactory mucosa lining of the nasal cavity, they are exposed to the environment and their lifespan is brief. Several reports say that OSNs are regularly regenerated during the entire life and that odorant environment affects the olfactory epithelium. However, little is known about the impact of the odorant environment on OSNs at the cellular level and more precisely in the context of early postnatal olfactory exposure. Here we exposed MOR23-green fluorescent protein (GFP) and M71-GFP mice to lyral or acetophenone, ligands for MOR23 or M71, respectively. Daily postnatal exposure to lyral induces plasticity in the population of OSNs expressing MOR23. Their density decreases after odorant exposure, whereas the amount of MOR23 mRNA and protein remain stable in the whole epithelium. Meanwhile, quantitative PCR indicates that each MOR23 neuron has higher levels of olfactory receptor transcripts and also expresses more CNGA2 and phosphodiesterase 1C, fundamental olfactory transduction pathway proteins. Transcript levels return to baseline after 4 weeks recovery. Patch-clamp recordings reveal that exposed MOR23 neurons respond to lyral with higher sensitivity and broader dynamic range while the responses' kinetics were faster. These effects are specific to the odorant-receptor pair lyral-MOR23: there was no effect of acetophenone on MOR23 neurons and no effect of acetophenone and lyral on the M71 population. Together, our results clearly demonstrate that OSNs undergo specific anatomical, molecular, and functional adaptation when chronically exposed to odorants in the early stage of life.
Keywords: development; electrophysiology; mice; molecular biology; olfaction; plasticity.
Figures



References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
