What the X has to do with it: differences in regulatory variability between the sexes in Drosophila simulans
- PMID: 24696400
- PMCID: PMC4007535
- DOI: 10.1093/gbe/evu060
What the X has to do with it: differences in regulatory variability between the sexes in Drosophila simulans
Abstract
The mechanistic basis of regulatory variation and the prevailing evolutionary forces shaping that variation are known to differ between sexes and between chromosomes. Regulatory variation of gene expression can be due to functional changes within a gene itself (cis) or in other genes elsewhere in the genome (trans). The evolutionary properties of cis mutations are expected to differ from mutations affecting gene expression in trans. We analyze allele-specific expression across a set of X substitution lines in intact adult Drosophila simulans to evaluate whether regulatory variation differs for cis and trans, for males and females, and for X-linked and autosomal genes. Regulatory variation is common (56% of genes), and patterns of variation within D. simulans are consistent with previous observations in Drosophila that there is more cis than trans variation within species (47% vs. 25%, respectively). The relationship between sex-bias and sex-limited variation is remarkably consistent across sexes. However, there are differences between cis and trans effects: cis variants show evidence of purifying selection in the sex toward which expression is biased, while trans variants do not. For female-biased genes, the X is depleted for trans variation in a manner consistent with a female-dominated selection regime on the X. Surprisingly, there is no evidence for depletion of trans variation for male-biased genes on X. This is evidence for regulatory feminization of the X, trans-acting factors controlling male-biased genes are more likely to be found on the autosomes than those controlling female-biased genes.
Keywords: Cis/trans gene regulation; X-chromosome; allele-specific expression; sex-biased expression.
Figures


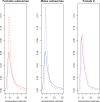
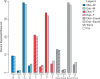
References
-
- Barton NH, Turelli M. Evolutionary quantitative genetics—how little do we know? Annu Rev Genet. 1989;23:337–370. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

